
Crossroads (iii) - a New Direction for Bioinformatics with Twelve Fundamental
This is the last post of a series of three commentaries. Previous ones can be seen at the following links.
Bioinformatics at a Crossroad Again - Which Way Next?
Crossroads (ii) - Is It Too Late to Acknowledge that Systems Biology and GWAS Failed?
The series started, when one of our readers asked about the remaining hard problems in bioinformatics. This reader is an expert in genome assembly, who contributed to development of a number of cutting-edge algorithms. The conventional answer would have been to suggest working on transcriptome assembly or metagenome assembly or developing more user-friendly version of Galaxy, or all of the above for big data, BIGGER data, BIGGEST DATA and never- ending iterations thereof.
However, we refrained from giving such an answer, because around the time the question came, we finished our electric fish paper and felt that a large part of contemporary bioinformaticians follow a false paradigm guided by unproductive motivations. We explained what we meant by ‘false paradigm’ in previous commentaries and will elaborate further here.
We arrived at above conclusion after closely following many bioinformatics papers, conference presentations, blog posts and discussions over the last few years. In our opinion, this false paradigm creates disconnect and conflicts between traditional biologists and those who are trying to analyze large genomic data sets. Examples of such conflicts can be seen in unflattering remarks at various places on ENCODE, GWAS, eQTL and other expensive misadventures.
-———————————————————————–
Entry of Computer Scientists into Biology
Influx of computer scientists into biology started in late 1990s, when the human genome project neared its end. Proliferation of inexpensive sequencing technology and other associated large-scale methods (microarray, chip-ChIP, etc.) created and continues to create such a need. We often speak with many of those researchers over email, and find them to be very smart, trying to bridge the gap between two distant fields.
The contemporary era has many similarities with late 1940s, when several physicists came into biology and brought in their expertise in X-ray crystallography and electron microscopy. Within a short period, that group established itself as the leaders of biological sciences. All physicists associated with Max Perutz in Cavendish lab and later ‘Laboratory of Molecular Biology’ made major breakthroughs in biological sciences. While reviewing the history of this era, we also noticed that their work brought together previously unconnected fields - biochemistry, genetics and evolutionary biology.
In contrast, the scientific contributions of various expensive bioinformatics- heavy projects of the current era had been marginal. ENCODE and modENCODE now take pride in their ability to collect data, but data collection was the trivial part of their original plan. Many expensive GWAS studies had been complete failures explaining only 2-3% of complex traits. It is unfortunate that this failure had been predicted by Ken Weiss in 2000, based on his previous experiences, but promoters of GWAS still do not pay attention to his arguments. Instead, to hide the failure, a recent GWAS study used statistical tricks and claimed to explain over 60% variations in height of all human beings. Ken Weiss has taken their argument to woodsheds in three excellent blog posts.
The height of folly: are the causes of stature numerous but at least ‘finite’?
Hiding behind technicalities to avoid having to change
Morgan’s insight–and Morgan’s restraint
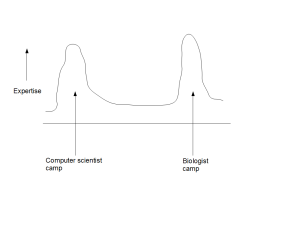
Speaking of general direction of the larger field of bioinformatics, what troubles us more is that there is a huge gap between the problems bioinformaticians consider as important and the problems biologists think as important. We went over the abstracts and motivation sections of many talks in WABI, a major bioinformatics conference (see WABIstan Report), and found them to be far away from explaining various aspects of life.
This is different from what the physicists did in 1950s, as we will explain in the following section.
-———————————————————————–
Differences between 1950s and 2000s
In 1945, renowned theoretical physicist Erwin Schrdinger wrote a book entitled ‘What is Life?’ and presented many basic questions in biology to physicists. Although the practicing biologists did not find anything novel in the book, it had profound influence on the physical scientists. It presented major questions in biology to physicists in their familiar languages.
Here is a good review of the book and its contribution to biology.
A few eminent scientists, such as Linus Pauling and Max Perutz, were critical of the contribution of What Is Life? PAULING (1987) wrote, When I first read this book, over 40 years ago, I was disappointed. It was, and still is, my opinion that Schrdinger made no contribution to our understanding of life.
…
In his introduction to the volume dedicated to Max Delbruck on his sixtieth birthday, STENT (1966) evaluated Schrdinger’s contribution to biology. He stated that it probably had little influence on professional biologists. But it had a great impact on physical scientists, who were only too happy to focus their intellect on a new and refreshing problem in the post-war years. Stent pointed out that there were significant gaps in Schrdinger’s genetic knowledge. He made no attempt to include chemistry in his discussion. Schrdinger’s knowledge was derived from a few published papers and earlier conversations with Delbruck. He conducted no experiments in genetics. His grasp of genetics was outdated. In What Is Life?, Schrdinger made no mention of the latest advance in genetics, namely the one geneone enzyme hypothesis that was supported by the discoveries of BEADLE and TATUM (1941), which ultimately contributed to molecular biology. He emphasized research on Drosophila even though by the 1940s genetics was taken over by those working with microorganisms, e.g., Neurospora. Even though it was Delbruck’s model that inspired Schrdinger’s interest in genetics, ironically Schrdinger did not seem to know that Delbruck had been working with bacterial viruses for the past five yearsleading to the phage school in genetics, which later provided the answers to many of the questions that Schrdinger raised in What Is Life?. The significant date was 1940, when Alfred D. Hershey, Salvador E. Luria, and Max Delbruck founded the phage group at Cold Spring Harborlong before Schrdinger was preparing his lectures in Dublin that eventually became What Is Life?.
In contrast, the computer scientists coming into biology in 1990s and 2000s were attracted by THE human genome project and various other large follow-up projects. After successfully constructing the human genome using the shotgun method, they advocated (i) inferring functions from the genome through GWAS, alignment and accelerated evolution, (ii) ‘shotgun biology’ (Brenner calls it ‘systems biology’), which attempts to collect various large noisy data sets and then infer functions by computational means. We see three fundamental flaws in the way computer scientists are proceeding -
(i) By trying to directly connect the genome and phenotypes in large complex organisms, they are skipping over an important intermediate modular block (see following section). A good analogy would be trying to explain how a large C/C++ code works by analyzing the characters in its code, when a better approach is to analyze the functions and classes within the code.
(ii) The reverse problem of shotgun biology is mathematically unsolvable in general, as Brenner argued in his paper ‘Sequences and Consequences’. We discussed this in our earlier commentary.
(iii) In terms of motivation, the bioinformaticians of current era are burdened by NIH mission of curing human diseases, whereas the physicists of previous era were trying to answer the fundamental question of ‘what is life’. It is ironic that the scientists of earlier era contributed more to improving human health by trying to answer questions in basic science, than scientists of current era with a direct mission to improve human health, but that is how basic science works. It is quite safe to argue that NIH’s channeling of money into various human health-related causes had been the most important contributor in reducing the quality of science in this country, but we will reserve that argument for another commentary. One relevant point is that by focusing on humans and humans only, NIH-funded large projects are increasingly moving away from evolution-based thinking. ‘Evolution’ is now packaged inside cute tree-building software programs.
“Nothing in biology makes sense except in the light of evolution” - Theodosius Dobzhansky, 1973.
-———————————————————————–
What is the solution?
In his paper ‘Sequences and Consequences’, Sydney Brenner not only showed the flaws of trying to infer functions from the genome, but also proposed an alternative based on the correct level abstraction - the cells. From the abstract -
We then solve the forward problem of computing the behaviour of the system from its components and their interactions. I propose that the correct level of abstraction is the cell and provide an outline of CELLMAP, a design for a system to organize biological information.
Digging further into his paper -
The genome must therefore form the kernel of any theory we construct but since transforming the information in a genome into the final living organism involves many complicated processes mediated by molecules specified in the genome, all of this will need to be known in considerable detail before we can read and understand genomes. There is no simple way to map organisms onto their genomes once they have reached a certain level of complexity. Thus while the genome sequence is central, it is a level of abstraction which is too cryptic to be used for the organization of data and the derivation of theoretical models. Proposals to base everything on the genome sequence by annotating it with additional data will only increase its opacity.
The correct level of abstraction is the cell. The cell is the fundamental unit of structure, function and organization of living systemssomething we have known for 180 years. This is the key feature of what I have called CELLMAP, a design for a biological information system that will allow us not only to handle the vast accumulation of data but also to generate and test hypotheses. CELLMAP is at once a map of the molecules within cells and a map of the cells in the organism; for microbes the cell is also the organism. All of us started as a single cell that multiplied to produce more cells, which differentiated into many different cell types to make up the tissues and organs responsible for our physiological functions. In choosing the level of the cell we avoid the question of whether our analyses should be top-down or bottom-up; instead, our approach is middle-out, because from the vantage point of the cell we can look down on the molecules that constitute it and look up at the organism that contains it. Furthermore, we can adopt a uniform conceptual architecture for all levels, viewing the organism as a network of interacting cells in the same way as we view the cell as a network of interacting molecules. As we shall see later, cell functions are generated by specific agglomerations of molecules just as physiological functions are exerted by specific collections of cells constituting our organs. In this way, our approach directly reflects the structure of biological systems and, as we reduce each level to the level beloworganisms to cells and cells to molecules we can then confidently complete the reductionist programme because the properties of molecules can be reduced to physics. This cannot be done in one step; we cannot decompose a human being into elementary particles and ask for the probability that these reassemble into the same human being, with the same genes, immune system and memories. This is absurd reductionism and if it could happen it would indeed be a miracle. Humans are not made in nature by the condensation of particle gases; as, is well known, each arises as a zygote produced by the fusion of two kinds of germ cells from the two parents. Interposed between quantum mechanics and a living organism are multiple levels of organization controlled by genes which have been generated by the processes of evolution, each step producing changes in the genetic material followed by natural selection of successful phenotypes. I was once accused by Rene Thom of being a constructivist, which I understand was worse than being called an empiricist; I replied that I took pride in it.
Based on our experience with the electric fish project, we believe ‘cellmap’ proposed by Brenner is the correct level of abstract, and moreover, making cells the center of attention will unify geneticists, biochemists, developmental biologists and bioinformaticians to solve common puzzles instead of alienating them from each other. We also believe evolutionary arguments can be correctly introduced by making the abstraction at the level of cells, whereas ‘genome evolution’ and phenotypic evolution are too distant to be connected. We discuss these in the following two sections.
-———————————————————————–
That is All Theoretical Talk. Show Me a Real Life Example of How to Use ‘Cellmap’
During early to mid-2000s, renowned developmental biologists Eric Davidson used to get invited at many bioinformatics conferences, until the computational scientists managed to bastardize his methods and then ignore him completely. These days, there are many ‘gene network’ packages, which are conceptually quite different (and inferior) from his gene regulatory networks. Similarly, his arguments about cis-regulatory control got overwritten by ‘epigenetics’, which these days mean methylation and histone modification, not Waddington’s classical epigenetics used by developmental biologists.
Those, who closely followed Davidson’s work over the years, find it match closely with Brenner’s description of cellmap. In fact, Davidson adds another dimension by investigating the build up of ‘cellmap’ from one single cell through execution of the developmental program coded in the genome. This is real science that will be remembered many centuries after ENCODE gets forgotten. We will briefly summarize the logical steps motivating his work so that you find it easy to follow his papers/books and understand why they are far more relevant to human biology than the hyped up genome wide association scams.
i) Human body constitutes of 150 or 200 different cell types, but they develop from one single cell.
ii) Unlike diabetes or height or intelligence or other traits GWAS-backers try to look for in the genome, the developmental program is very likely to be coded in the genome. So, unless we can explain why our left hands always look like right hands, it is futile to understand why person X is taller than person Y.
iii) Speaking of predictable developmental program, the early development of almost all animals follow very similar stages of formation of ectoderm, mesoderm and endoderm. Various derived cell types form from one of those three ‘layers’.
iv) So, Davidson uses his genetic tools to figure out how the ectoderm, mesoderm and endoderm form from the single embryonic cell. He picks sea urchin, because it is the simplest organism with early developmental patterns similar to humans (deuterostome).
v) Points i-iv are common knowledge among all developmental biologists, but unique aspect (at least when he started) of Davidson’s work was in connecting the developmental program with the genome in the following way. He divided all genes into two categories - informational genes and effector genes. The informational genes, such as transcription factors, tell a cell to be neuron or liver cell, but being in certain collective state. For argument’s sake, let us assume that a genome has 200 informational genes and they can be in 0 or 1 state. When the first 100 of them are in 1 state and the remaining 100 are in 0 state, a generic cell turns into a neuron. Developmental program works through progression of state changes of those informational genes, and Davidson tracked them using his ‘gene regulatory network’.
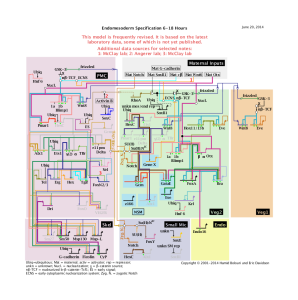
The above image, taken from Davidson’s page, shows the dynamics of development of Brenner’s ‘cellmap’.
vi) Here is how things can get really cool. If similar gene regulatory networks (or state specification map of informational genes’ are constructed for many different organisms, one would start to see conserved patterns, just like we find conserved patterns by comparing genomes. However, the conserved loops in gene regulatory network can be directly connected to phenotypes in the form of developmental patterning. You can check this paper by Veronica Hinman and its references to find examples of how to connect genome, cellmap and body patterns.
-———————————————————————–
A Short Summary of our Electric Fish Experience
We first posted Brenner’s article in 2012 (see Thought-provoking Article on Systems Biology from Sydney Brenner), but did not fully appreciate the wisdom in it until going through the exercise of electric eel genome project. Therefore, we will summarize our experiences with the electic eel project here.
At first, we started with sequencing and assembling the genome of electric eel, but we were very reluctant to publish another genome paper of the form (“X is a cool organism. Here is its genome.”).
It is very difficult to say anything biological from the genome, and most genome analysis projects employ three methods - (i) types of genes not present in the genome (such as electric eel genome lacks many vision-related genes), (ii) gene classes present in large number (receptors in various insect genomes), (iii) genes undergoing accelerated evolution. However, none of these methods are good for explaining important phenotypes. Sure, the electric eel lives in mud and does not use its eyes, but that says very little about electric organs.
Given that there is another far more convenient method to connect genes with phenotypes, we saw no reason to stick to the genome only. We realized the power offered by de novo assembled transcriptomes very early, and did not see any reason to stick with the genomes. For reference, check -
De Novo Transcriptome Assemblers Oases, Trinity, etc.
De Novo Transcriptome Assemblers Oases, Trinity, etc. II
De Novo Transcriptome Assemblers Oases, Trinity, etc. III
De Novo Transcriptome Assemblers Oases, Trinity, etc. IV
Once the focus moved away from the genome to transcriptome, the questions being asked got more targeted to the biological observations. Why not compare gene expression in electric organs of electric eel with other south American electric fish species? Why not compare transcriptomes of different electric fish, which independent evolved electric organs?
By trying to explain the behavior of a particular cell type (electrocyte) instead of ‘understanding the genome’, we believe we made far more contribution to biology than a large number of recent genome project. A more conventional approach would have been to sequence the genomes of many different kinds of electric fish, publish a ‘high-profile’ comparative genome paper and get rebuked by Dan Graur.
-———————————————————————–
Twelve Fundamental Problems
Now that we take our center of attention away from genomes to cells, what are the fundamental problems to solve? We believe thinking in terms of fundamental problems is far more productive than being motivated to cure human diseases.
We came across an article by Mike White (whose scientific work was mentioned earlier in our blog) telling us that no fundamental problem is left unsolved in biology.
I’ve heard a senior colleague say that there is nothing fundamental left to be discovered in biology. It’s a nagging worry some people have, including myself. What’s left, according to some (including one of molecular biology’s founders Sydney Brenner), is to work out the details of particular systems, implied by already established paradigms - kind like chemistry.
Given that he quoted heavyweights like Brenner (although without reference), we had to give weight to his argument. So, we went through a large number of relevant papers and books on evolutionary biology (some listed in following section), but came to the conclusion that the fundamental landscape is as open as ever before. A somewhat arbitrary list of the most important open questions in biology are given below. We will list most of them without description, briefly explain a few and elaborate further on all of them in future blog posts.
Bioinformaticians have an edge over other biologists in attempting to answer these questions, because they can seamlessly move from one organism to another in their computer. However, unless someone asks the fundamental questions, he does not get to answer any.
i) Prokaryote-eukaryotic discontinuity
Evolutionary theory does not like discontinuities, yet we see a large unexplained discontinuity between the prokaryotic cells and eukaryotic cells. How the complex eukaryotic cells evolved from prokaryotic cells is quite unclear even today and genome sequencing of bacteria and archaea added to the dilemma instead of offering a straightforward answer. Dan Graur posted an informative commentary on the topic, and it is a section of his upcoming book.
We consider this as the most important fundamental question in biology, because to work with a ‘cellmap’ in eukaryotic multicellular organism, it will be important define the cell. Given that many genes supposed to be present in complex multicellular organisms are found in sponges, ctenophores and even unicellular chaonoflagelletes, the understanding of eukaryotic cells will be more and more important.
ii) Evolution of eukaryotic cilia
About nine years back, we worked on flagella in Chlamydomonas and was fascinated by intraflagellar transport.
Intraflagellar transport or IFT is a bidirectional motility along axonemal microtubules that is essential for the formation (ciliogenesis) and maintenance of most eukaryotic cilia and flagella.[1] It is thought to be required to build all cilia that assemble within a membrane projection from the cell surface. Plasmodium falciparum cilia and the sperm flagella of Drosophila are examples of cilia that assemble in the cytoplasm and do not require IFT. The process of IFT involves movement of large protein complexes called IFT particles or trains from the cell body to the ciliary tip and followed by their return to the cell body. The outward or anterograde movement is powered by kinesin-2 while the inward or retrograde movement is powered by cytoplasmic dynein 2/1b. The IFT particles are composed of about 20 proteins organized in two subcomplexes called complex A and B.[2]
IFT was first reported in 1993 by graduate student Keith Kozminski while working in the lab of Dr. Joel Rosenbaum at Yale University.[3][4] The process of IFT has been best characterized in the biflagellate alga Chlamydomonas reinhardtii as well as the sensory cilia of the nematode Caenorhabditis elegans.[5]
It has been suggested based on localization studies that IFT proteins also function outside of cilia.[6]
The related genes are highly conserved all the way from chlamy to human, which means they play important roles. How this complex apparatus evolved in the first place is unclear.
iii) Origin of life - RNA world
What happened before DNA–>RNA–>protein? How did ribosome evolve? These questions are far from answered.
iv) Origin of sex in eukaryotes
This is one topic, where bioinformaticians can contribute strongly by developing algorithms to identify ‘sex chromosomes’ (often far short of entire chromosomes) in the newly sequenced genomes.
v) Neuro-muscular junction and role of electricity
vi) Flexibility of genome
Once you look down from the cells to the genome, some of the statistically divergent genomes appear as odd. How do highly polymorphic genomes manage to produce very stable phenotypes? How much can a genome be randomized without any change in biological properties?
vii) Animal body plan
Given that everything in the genome moves around, why do the Hox genes have to always stay together?
viii) Evolution of receptors
ix) Death and Aging
In early 2000, a number of large-scale experiments came out in yeast, but one of them stood out from the others in terms of interpretation. It was the yeast gene deletion experiment. Individual deletion of certain genes from yeast genome resulted in death of yeast cells, but how does one interpret ‘death’ as a condition? What does it mean in terms of network? To understand the dying process of a cell, we did an interesting experiment. The paper took a different turn, but the larger topic is still open. Nick Lane’s book cited below has an interesting section covering this topic.
x) Evolution of immune systems in eukaryotes
xi) Evolution of Metabolic Network and Photosynthesis
xii) Brain, intelligence, Consciousness (and maybe Eusociality in bees/ants)
-———————————————————————–
Influences
We determined the ‘fundamental problems’ based on reading a large number of books and discussions with various scientists. To start with, we read older books on evolution by Charles Darwin and Stephen J. Gould, and also the papers of Fisher, Wright and Haldane related to derivation of ‘modern synthesis’. In addition, we read many foundational papers in evo-devo. The following books/paper were helpful.
Ken Weiss and Anne Buchanan - Genetics and the logic of evolution
Eric Davidson - The Regulatory genome
Bruce Alberts - Molecular Biology of the Cell
Dan Graur, Chris Amemiya and Eric Davidson helped me by patiently answering many simple questions or guiding me to appropriate references.
-———————————————————————–
Please give your feedback
We would like to get your feedback on the above commentary, because starting from next year, we like to take the discussions in this blog along the direction of those twelve fundamental problems and any other important problem suggested by others. We believe it would bring down the distance between bioinformatician and biologist camps, and let everyone collectively answer major questions related to living systems.