
Rectangular Graph Algorithm
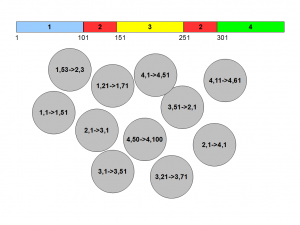
Let us say your genome looks like this -

Two red blocks are identical, but blue, yellow and green segments are all different. Also, we know the pieces and their distances, but we do not have them assembled as nicely as above. How do we assemble them?
Let us try one strategy, where we start picking pairs of positions 50 nucleotide apart and form circles. In most cases, both ends of pairs will fall on the same colored segments, but in some cases, they will be fall on different segments. We also know that ‘1,1->1,51’ will be followed by ‘1,2->1,52’ and so on. So, we have some way of connecting the pieces in a long chain, but there are too many circles.
A better strategy is to create small rectangles with two edges being formed by two segments of the genome. Those two segments can be the same, in which case, the rectangle is a square. We also draw a line that represents nucleotides 50 bases apart. Essentially, we have the same problem represented with fewer pieces.
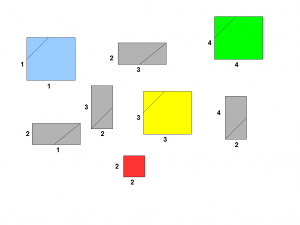
We need to connect the rectangles together so that their edges match properly, and voila, you have the genome assembled.
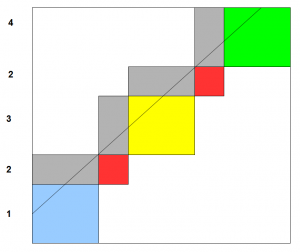
How to connect the rectangles? Essentially you form a graph based on similar edges of those rectangles and find a path through the graph. We will explain that later, but the answer is already known to the Chinese postman, who does not want to walk and the mathematician, who wanted to cross bridges.
-—————–
Homework for readers (except those from France) -
1. What happens, if the second red block is reverse complement of first?
2. Can we reconstruct the genome, if 2 is missing?