
Genome of a 'Living Fossil'
Imagine you got a chance to enter the Jurassic park, collect DNA samples from a T. rex and come back home to sequence its genome. A recent genome paper from Chris Amemiya and colleagues achieved similar feat even without including Stephen Spielberg on the authors’ list.
The above description is a bit exaggerated, because the African fish they sequenced is not really extinct like dinosaurs. However, coelacanth fish is so rare that it was believed to have gone extinct 70 million years back, and the first sighting in 1938 caused worldwide sensation. Since then, only 309 individuals have been recorded according to the Nature paper.

Coelacanth fishes fascinate biologists for another reason. They look
exactly identical very similar (check comment section) to their
fossil records dating back 300 million years, and likely have evolved slowly
over all these years. Therefore, they are indeed ‘living fossils’ representing
an ancient era, and their genomes can provide clues about how fishes diverged
from tetrapods.
The genome was sequenced at Broad Institute using Illumina sequencing, and assembled using ALLPATHS-LG. Annotation was done using both ENSEMBL and MAKER pipelines. In the following description, 40 kb ‘FOSSILL’ represents paired end Fosmid junctions (Williams, L. J. et al. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res, doi:gr.138925.112).
The Latimeria chalumnae assembly, LatCha 1.0 was constructed from 180 bp paired end fragment libraries (61X coverage), 3 kb jumping libraries (88X coverage), and 40 kb FOSSILLs (1X coverage). All libraries were sequenced by Hi-Seq Illumina machines, producing 101 bp reads.
Genome analysis of coelacanth showed that the protein-coding genes were evolving very slowly, as expected. The paper also did quite a bit of evolutionary comparison between fishes and tetrapod, and found that lungfish, and not the coelacanth, was the closest living relative of tetrapods. In fact, establishing the above point could not have been possible without NGS. Lungfish genome itself is extremely large (50-100 Gb), but with RNAseq, it was possible to get its protein-coding genes without doing a full genome sequencing.
The paper had very interesting results on Hox cluster, and especially a conserved noncoding enhancer (CNE) near Hoxa14. The CNE region is present in tetrapods even though Hoxa14 is not. This is related to previous work from Dr. Amemiya’s previous work on Hox genes, which he published in 2010 (Amemiya, C. T. et al. Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome.Proc. Natl Acad. Sci. USA 107, 36223627.).
The supplementary section is also full of gems. We wish some observations from analysis on lncRNA made into the main paper.
-——————
Here is timeline of the project based on Dr. Amemiya’s comment below. Two large gaps represent getting initial NIH approval for sequencing and getting updated approval for Sanger–>NGS switch.
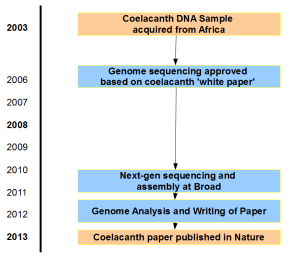
Original whitepaper submitted in 2006 is available here.
-—————–
NY Times coverage with comments from authors of the paper -
Fishs DNA May Explain How Fins Turned to Feet