
SOAPdenovo2 Demystified - part 1
It was lot of work parsing through 40,000 lines of completely unknown code, but we believe we finally figured out most of SOAPdenovo2 genome assembler. That does not mean we have become an expert of any sort, but at least we have good understanding of logical flow of the code to work through all details. In the next few commentaries, we will go through various parts of the code, explain their functionality and algorithms here, and also update the corresponding wiki pages.
Before we begin, here is an updated figure explaining the roles of 50 C files in the ‘standardPregraph’ subdirectory of SOAPdenovo2. You will see some changes from our previous discussion. Even the following figure is ‘work in progress’.
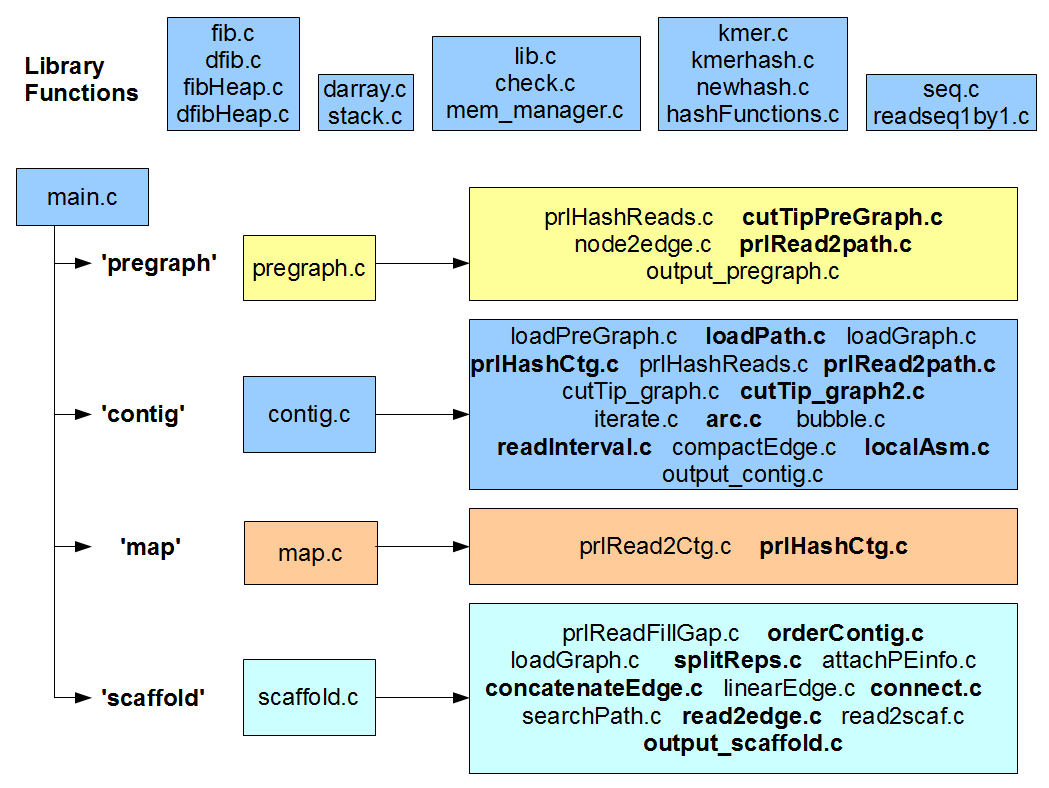
How did we proceed to understand the code? We have some understanding of de Bruijn graph-based assembly algorithms, and we also looked into the codes of few other assemblers. With that knowledge, we decided to dig into SOAPdenovo2 code. Still, it is very difficult to process ~40,000 lines of C code without killing oneself, but one can simplify the task by looking into various functions. At first, we parsed the code into functions, and created separate wiki pages for each file listing all its functions (e.g. wiki page for functions of OrderContig.c). Even that was not enough, because we had 750 functions to look at. So, we decided to rank functions based on how often they were used and checked the functions with the highest rankings. That is the bottom-up approach for understanding complex code. Separately, we started from ‘main’ program and continued to check the functions in top-down manner. Somewhere down (or up) the road, those two methods met each other and things started to make sense.
Anyway, all those are unimportant now that we know which function does what task. Let us dig into the code together. Our discussion will be divided in the following manner.
a) We will discuss the ‘library’ functions and their associated algorithms. You will notice that similar ‘standard’ blocks are used in many other bioinformatics programs (including aligners) and some non-bioinformatics programs as well.
b) We will go through the threading related codes. Files with names starting with ‘prl’ (prlHashCtg.c, prlHashReads.c, prlRead2Ctg.c, prlRead2path.c, prlReadFillGap.c) parallelize their operations through multiple threads, and they have similar format.
c) We will explain the data structures used in SOAPdenovo code. Those data structures are defined in header files stored in ‘inc’ subdirectory. Please start with extvab.h and global.h, if you want to explore yourself.
d-g) We will go through the codes related to ‘pregraph’, ‘contig’, ‘map’ and ‘scaffold’ modules of SOAPdenovo2.
h) We will discuss the sparsePregraph module available from sparsePregraph directory.