
Top Ten Genomes – (vi) Oxytricha trifallax for MIC and MAC in Ciliates
Last year, Laura Landweber’s lab published the sequence of germline genome of Oxytricha trifallax.
For anybody interested in genome sequences, this one completely blows one’s mind.
Highlights
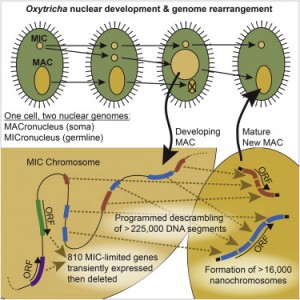
Oxytrichas encrypted germline genome contains >3,500 scrambled genes
225,000 DNA segments assemble, some combinatorially, to form the somatic genome
Thousands of gene segments for different somatic loci interweave with each other
Hundreds of germline-limited genes are expressed during development, then deleted
Summary
Programmed DNA rearrangements in the single-celled eukaryote Oxytricha trifallax completely rewire its germline into a somatic nucleus during development. This elaborate, RNA-mediated pathway eliminates noncoding DNA sequences that interrupt gene loci and reorganizes the remaining fragments by inversions and permutations to produce functional genes. Here, we report the Oxytricha germline genome and compare it to the somatic genome to present a global view of its massive scale of genome rearrangements. The remarkably encrypted genome architecture contains >3,500 scrambled genes, as well as >800 predicted germline-limited genes expressed, and some posttranslationally modified, during genome rearrangements. Gene segments for different somatic loci often interweave with each other. Single gene segments can contribute to multiple, distinct somatic loci. Terminal precursor segments from neighboring somatic loci map extremely close to each other, often overlapping. This genome assembly provides a draft of a scrambled genome and a powerful model for studies of genome rearrangement.
Or check some of the reports in popular press -
Single Cell Smashes, Rebuilds Its Own Genome
The pond-dwelling, single-celled organism Oxytricha trifallax has the remarkable ability to break its own DNA into nearly a quarter-million pieces and rapidly reassemble those pieces when it’s time to mate, the researchers report in the journal Cell. The organism internally stores its genome as thousands of scrambled, encrypted gene pieces. Upon mating with another of its kind, the organism rummages through these jumbled genes and DNA segments to piece together more than 225,000 tiny strands of DNA. This all happens in about 60 hours.
This Bizarre Organism Builds Itself a New Genome Every Time It Has Sex
Unlike humans and most other organisms on Earth, Oxytricha doesnt have sex to increase its numbers. It has sex to reinvent itself.
When its food is plentiful, Oxytricha reproduces by making imperfect clones of itself, much like a new plant can grow from a cutting. If theyre well fed, they wont mate, said Laura Landweber, a molecular biologist at Princeton University and lead author of a recent study on Oxytricha genetics. But when Oxytricha gets hungry or stressed, it goes looking for sex.
When two cells come together (as in the image above), the ultimate result is: two cells. Theyve perfected the art of sex without reproduction, Landweber said. The exterior of the two cells remains, but each cell swaps half of its genome with the other. Theyre entering into this pact where each one is going to be 50 percent transformed, Landweber said. They emerge with a rejuvenated genome.
For those interested in learning about scrambled genomes in ciliates, the following website is helpful.
Ciliated protozoans (Phylum Ciliophora) are characterized by the presence of cilia used for locomotion, and the presence of two types of nuclei: a somatic nucleus macronucleus (MAC) which provides templates for the transcription of all genes required for vegetative growth, and a genetic nucleus micronucleus (MIC) used for the exchange of meiotic products during sexual reproduction.
Following conjugation (sexual reproduction), during which haploid gametic nuclei are swapped between pairs of mating cells and a diploid zygotic nucleus formed, new MIC and MAC are generated from copies of the zygotic nucleus. DNA in the MIC remains organized in the form typical of most eukaryotes, with pairs of large chromosomes. In contrast, chromosomes in the MAC genome undergo extensive DNA fragmentation, elimination and amplification, followed by telomere addition (Prescott 1994), resulting in many small acentric chromosomes.
The extent of MAC genome reorganization varies greatly among ciliate species. In ciliates belonging to the class Spirotrichea the level of DNA processing in the formation of a new MAC is extraordinary: the original zygotic chromosomes are fragmented at tens of thousands of positions, 95% of the DNA complexity is lost, and the resulting chromosomes sometimes referred to as “nanochromosomes” are amplified to thousands of copies each (Prescott 1994). In spirotrichs, each MAC chromosome typically contains a single gene flanked at the 5’ and 3’ ends by very short (<150 nt) untranslated regions and telomeres (Prescott and Dizick 2000; Cavalcanti et al. 2004a). The size of these molecules ranges from 0.25 kb to 35kb, and each is present at an average of 1000 copies.