
Highlights of Gene Myers' Talk at #AGBT15
We covered Myers’ talk at AGBT last year (Dazzler Assembler for PacBio Reads Gene Myers), where he announced dazzler assembler. This time you can view his talk through live-streaming, thanks to Pacbio.
His this year’s first talk has two major highlights.
1. Perfect assembly
Myers argued that perfect assembly is possible as long as the average read length is larger than the longest repeat. To quote him - “repeats less than 13kb is solved in pacbio. 13kb from median reads length minus 2 franking lengths.” Error rate is not a factor.

In this context, readers are encouraged to take a look at various papers (e.g. Bresler et al. and more recent Shomorony et al.) from David Tse’s group at Berkeley.
2. Release of DAscrubber
This year, he is planning to release the second module of DAZZLER pipeline and it will be used to clean reads. Here is his slide on various existing assembly pipelines.
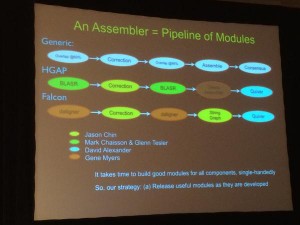
Regarding the Daligner released last year, he said “I pulled every trick I ever learned” with Daligner to get 25x - 40x speed improvement over BLASR”.
We will add to this post after his tomorrow’s talk.
-——————————————–
The second talk of Gene Myers is described the DAscrubber module, and proposed a new and efficient standard for storing alignment data instead of bam format.
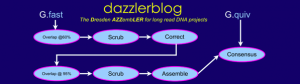
The purpose of this step is to clean the reads before finishing the final assembly.
On what DA scrub does
“DAscrub “TracePoint” really cleans up data; removes hairpins; patches gaps; finds chimeras; etc”
“Scrubber favors editing rather than not editing”
“using intrinsic QV (not from the instrument, but from other reads)”
“99% reads are now perfect; double-scribing increases this to 99.99%”.
On rejection of bam format
“encoding the edit script (BAM) is hugely space inefficient but computing alignment as needed is hugely time inefficient”
“new standard of sequencing file : trace of each point + difference between successive points. Space & time efficient” [as opposed to bam files]
“Keep distance between points, keep # of differences, both space and time efficient, encoding alignments. 60x faster”
“describes the ideas of “Trace point” to compress alignment data for efficiency, allowing reconstruction alignment in linear time”