
Explosion of Transcriptome Data on Drosophila
While reviewing our GEO Trends section on different model organisms, we noticed an explosion of submitted experiments on Drosophila (fruit fly) over the last three years.
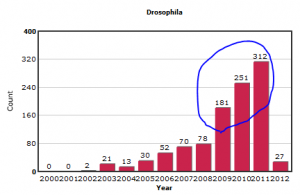
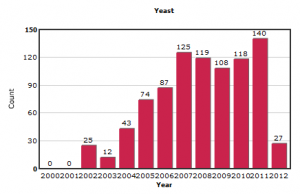
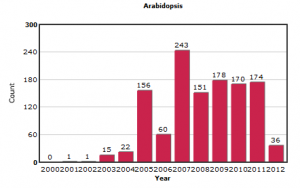
In contrast, the submissions on other model organisms like yeast and Arabidopsis appear more stable.
We first expected the new submissions to be from NGS sequencing, but after going through the data, we found out that much of the new data came from one platform.
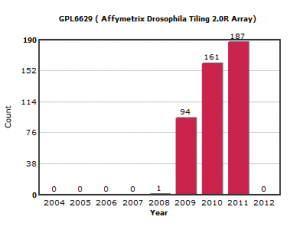
GPL6629 is a tiling array platform for Drosophila genome developed by Affymetrix. It tiles one strand of the entire Drosophila genome with 25-mer probes at a resolution of one probe/38 nucleotides. We started using it in one of our experiments, but the PI of the project made a smart decision and decided to go for ChIP-seq. After a lot of analysis and comparisons, we concluded that ChIP-seq provided better resolution than ChIP-chip. You can read the rest about that work here -
[High resolution mapping of Twist to DNA in Drosophila embryos: Efficient functional analysis and evolutionary conservation -
Anil Ozdemir, Katherine I. Fisher-Aylor, Shirley Pepke, Manoj Samanta, Leslie Dunipace, Kenneth McCue, Lucy Zeng, Nobuo Ogawa, Barbara J. Wold, and Angelike Stathopoulos.
That accounts for only one experiment out of so many others submitted on Drosophila. Where did the rest come from?
We looked into the details of submissions on GPL6629 and found over 80% to come from the group of Susan Celniker at Ontario Institute for Cancer Research as part of a huge modEncode project. The goal of the project from GEO website -
We will generate over 600 RNA samples in biological triplicate and use them to generate expression profile maps detailing the sites of transcription across the fly genome using whole genome tiling arrays at 38 bp resolution as a broad survey of the transcriptome and 7 bp arrays resolution to identify at high resolution transcripts ends, splice sites, and small RNAs.
Their paper was published in Nature last year ( Graveley BR, Brooks AN, Carlson JW, Duff MO et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011 Mar 24;471(7339). ), but I expect this volume of data to result in many interesting discoveries for years to come.
The second largest contributor to explosion of Drosophila transcriptome data was Kevin White’s group at University of Chicago. Their group had been working on Drosophila tiling arrays for long time and was the first to publish tiling array map of Drosophila genome.
This commentary gives you a bird’s eye view of what is going on in fruit-fly land. In future, we will go into the details of these experiments and discuss more about what they found.