
What Fraction of Genome is Really Functional? (Our lncRNA story)
ENCODE’s bold claim that 80% of human genome is functional created quite a bit of controversy, but also got everyone distracted from the real question - what fraction of non-coding transcripts are functional? This commentary will not answer that question, but at least present a method developed by us in 2006 that will get researchers one step closer than where they are today. In 2005-2007, we started answering the question of functionality of non-coding RNAs systematically, but our efforts got derailed by four major non-scientific shortcomings of our approach. We will get to those in a follow-up commentary.
Today I called up after long time a researcher with whom we collaborated in 2006, and realized how much our perspectives have changed. In 2006, he was a post-doctoral scientist at UCSF. I convinced him that the genomes were full of additional transcripts beyond what protein-coding genes covered and signed him up for our interesting project. I told him - “Forget human genome. Even in yeast genome, I can show you many non-coding transcripts and their presence makes your traditional understanding based on protein- coding genes invalid.” My suggestion came as a complete surprise to him, because everyone in their lab assumed yeast genome to be fully annotated.
If you think about it, that is the ENCODE message minus hype. Between 2004 and 2007, I told any biologist who wanted to listen about the presence of mystery RNAs all over the genomes. I gave talks at all kinds of places saying that the genome annotation pipelines were incomplete, because they only identified the protein-coding genes. In almost all my papers, I tried to add at least one section discussing mystery RNAs (now known as lncRNAs). How did I know about them? Between 2003 and 2007, we worked on a technology that comprehensively measured all regions of the genome for signs of transcription. Our first paper was on human genome and we identified a number of additional transcribed regions. That was not a surprise in 2004, because human genome was only 3 years old and was expected to be poorly annotated due to its size.
Global Identification of Human Transcribed Sequences with Genome Tiling Arrays
However, we saw the same pattern in Arabidopsis, which was relatively well annotated and had shorter introns. Our Arabidopsis paper did not mention non-coding RNA, because I could not convince our biologist collaborator that they were not artifacts. At first I gave him the same treatment that Ewan Birney offers to Dan Graur - ‘the guy did not catch up with advanced technology and remains stuck in 70s or 80s’. I soon got humbled, when Dave Nelson, then a brilliant PhD student in our collaborator’s lab, ran few Northerns on the most convincing-looking noncoding RNAs in Arabidopsis and got nothing. That was very puzzling, and we worked through the details to figure out that those noncoding transcripts were indeed artifacts generated by standard oligonucleotide array protocols. It was a fun paper even though we did not find what we looked for.
That only removed a subset of non-mystery RNAs and many others still remained. Some did not look like artifacts, because they had identical intron-exon boundaries in human and mouse data, and I was dead sure that they were real. There was only one problem. No serious biologist believed they were real until we showed their functions and nobody wanted to do biochemical experiments until we made a convincing case that they were real.
After many long hours of thinking and discussing, my colleague Victor and I stumbled upon an interesting idea. Let me explain it with an analogy first (to get dear reader Istvan upset) and then present the real biological approach.
The popular image of New York city consists of big shot bankers and other glamorous people, but are they functionally the most important? There is an easy way to find out and people of New York did so in 1981. In that fateful year, garbage collectors of Big Apple decided to stay home for 17 days. Very soon trashes piled up all over the place and NY City residents became aware of invisible but functionally important people.

The above analogy is an imperfect description of what we did. We decided to turn off a major RNA processing pathway (RPP1) and check which noncoding transcripts started to pile up. We argued that if a noncoding RNA was doing useful things in the cell, it was likely getting processed by one or other RNA processing complexes. Therefore, by turning off the processing pathway, we would see piling up of large amount of unprocessed transcripts (or ‘trash’ if you call it). The method worked. When we depleted RPP1, an essential RNA- processing gene, many regions of the yeast genome lighted up.
What followed was bioinformaticians’ paradise. I sat with 100 or so reasonably convincing noncoding RNAs in one of the most well-studied genomes, and turned every stone to say something interesting about them without doing wet lab experiment. We played with sequence conservation among yeasts, RNA folding, promoter sequence and every possible trick, but did not get anywhere soon. The reason was simple. If we could show some real functions for few of those noncoding RNAs, we could tell others that many other noncoding sequences identified by expression studies were real. Experience showed us that finding some real function was more important than talking about wonders of technology, which we did for long enough by that time.
Finally, I struck gold in an old MCB paper from 1993. The authors did very thorough characterization of DRS2 gene and found it to be associated with two wildly different functions. That itself is nothing unusual, because many distinct pathways can get connected through unknown mechanisms. What got me interested was an experiment by the authors. They found one function to go away when they cut 3’ half of the gene, but stayed, when they mutated the 5’ half of the gene. Our mystery noncoding RNA was antisense to the 3’ half of the gene. We could fully explain his experiments by associating one of the functions with DRS2 and the other one with the antisense transcript.
I showed the paper to Victor and he immediately jumped up. The evidence was lying right in front of everyone’s eyes, as you can see below. The authors submitted this gel image in their paper, and lo and behold, a large spot with the right size of the antisense RNA was right there in their gel, but they did not discuss it in their paper.
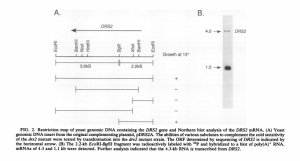
Victor called up the senior author of the paper, and we found out that he saved his RNA materials from the study twelve years back. It was a miracle, because we were not ready to do biochemistry in the lab. For the next few days, we waited in suspense. The materials arrived, we sent it for sequencing and found the antisense RNA to be exactly where array data showed it to be present (check second figure below).

If you are still with me, let me recap what we got at this point.
1. For several years prior to that time, we saw long mystery RNAs all over tiling array data, but could not convince anyone that they were real.
2. Our expression study of turning off RNA-processing pathway added with tiling array showed that those 100 or so of shortlisted transcripts not only had signatures of expression (as in 1) but were also differentially expressed. Their accumulation suggested that they were at least processed by the RNA- processing complex. –> adds one more layer of credibility.
3. Although we did not get our hands dirty with biochemical characterization, Ripmaster et al.’s 1993 paper did it and found one of those mystery RNAs to be indeed functional. They did not interpret their results that way, because they were following DNA–>RNA–>protein model and were not aware of noncoding RNAs.
If 1, 2 and 3 were correct, the implications were that at least some fraction of mystery RNAs were real in yeast. If they were real in yeast, they were likely to be real in human, mouse, fruit fly, honey bee and every other organism we touched with our tiling array experiments (after accounting for artifacts, etc.). I thought that was a big deal. It seemed to have become big deal 6 years later, when biologists ‘discovered’ long noncoding RNAs.
I called up a Science editor to explain why our results were important. After some discussion, she asked whether we discovered new miRNAs. Editors of glam journals go by buzzwords and miRNA was the buzzword of the day in 2006. You have anything more interesting than miRNAs? Come back after 5 years and resubmit, when your field gets hot. We called up an old and well-respected RNA biologist (Victor’s PhD adviser), who was more receptive to our ideas. His lab conducted full biochemical analysis and confirmed the function of RNA antisense to DRS2. Like us, he was also not in hype business and reported exactly what he got in RNA journal.
When I read papers on long noncoding RNAs, we find them stuck at the same place as we were in 2006. I still think turning off of various RNA processing pathways is the ideal way to proceed, and the researchers need to go up from simpler organism to complex instead of all focusing on human. Sure human genome is important, but so is evolution.
Follow up commentary will explain why our paper got very few citations despite being one of the earliest (three years before John Rinn for sure) in presenting functionality of what is now known as long noncoding RNAs. If you are a young graduate student, do not make the mistakes that we did.