Note: These tutorials are incomplete. More complete versions are being made available for our members. Sign up for free.

Other Evidences - RNAseq
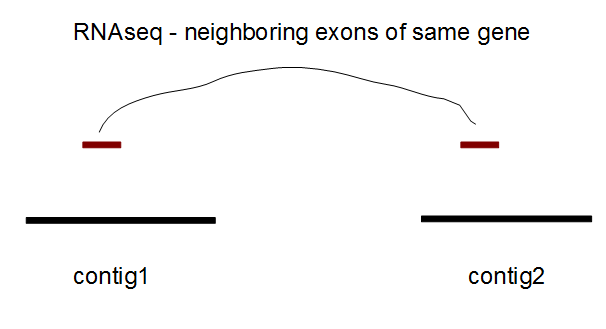
RNAseq data can also be used to improve the scaffolding of an assembled genome. The procedure is demonstrated in the above image. If two consecutive exons of a gene assembled from RNAseq are found to be located in two different contigs or scaffolds, it is likely that those scaffolds are in neighboring regions of the same chromosome. Unlike in paired end reads, the actual distance between the contigs is not possible to determine from RNAseq, but an easy solution is to estimate it from the average distance between neighboring exons in genes already mapped to the same scaffold. Some caution is warranted, because the leading 5’ exon in some mammalian genes are located far away in the chromosome from the remaining exons.
