Note: These tutorials are incomplete. More complete versions are being made available for our members. Sign up for free.

Illumina Reads
Relative orientation and directionality of NGS reads in paired end or mate pair libraries is an important factor to keep in mind, while assembling them into larger contigs. Most people working on NGS data understand that reads in paired end libraries come from opposite strands, but forget to take into account that the reads can be inward or outward looking. Let me elaborate.
This morning, we were working on an Illumina short read library, and when we mapped the reads on to the genome, we got the following locations -
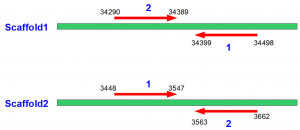
@AAA-BB111_0123:1:1:711:111211#TNNNNN/1 – SCAFFOLD1 34399 @AAA-BB111_0123:1:1:711:111211#TNNNNN/2 + SCAFFOLD1 34290
@AAA-BB111_0123:1:1:715:11330#TNNNNN/1 + SCAFFOLD2 3448 @AAA-BB111_0123:1:1:715:11330#TNNNNN/2 – SCAFFOLD2 3563
etc.
Here is a second library we were working on. This one is mate paired with longer distance between the ends of reads.
@AAA-BB111_0022:2:1101:8837:1317 1:N:0: – SCAFFOLD21 3264 @AAA-BB111_0022:2:1101:8837:1317 2:N:0: + SCAFFOLD21 6067
@AAA-BB111_0022:2:1101:8837:1318 1:N:0: + SCAFFOLD22 89142 @AAA-BB111_0022:2:1101:8837:1318 2:N:0: – SCAFFOLD22 86945
etc.
To understand the difference between two libraries, let us plot the genomic locations of the reads.
Here is the first library -
and here is the second -
In the above figures, arrows go from 5′ to 3′ direction.
You can immediately recognize the difference between two libraries. In the first case, the arrows are directed inward, whereas in the second case they are directed outward. Moreover, this directionality is independent of whether the read from library 1 is on W strand and read from library is on C strand, or vice versa. Directionality is an intrinsic property of the library itself, and decided on how the sequencing was done.
The above information becomes important, when we try to assemble the reads based on pairing information. Suppose the read 1 falls on scaffold3222 and read 2 falls on scaffold2162. This suggests that scaffold3222 and scaffold2162 are close to each other, but whether scaffold3222 is on the 5′ side of scaffold2162 or on the 3′ side depends resolving the directionality information correctly.
Here is a simple pre-processing rule that can help one sort out all strand-related complexities, while aligning or assembling reads from paired Illumina libraries. Suppose you have 2 short read libraries name A/1 and A/2.
If they are inwardly directed, rewrite entire library A/2 by taking reverse complement of all reads. When you do that, each read pair from A/1 and A/2 will fall on the same strand with A/2 following (i.e. on the 3′ side of) A/1.
If they are outwardly directed, rewrite entire library A/1 by taking reverse complement of all reads. When you do that, each read pair from A/1 and A/2 will fall on the same strand with A/2 following (i.e. on the 3′ side of) A/1.
Please remember that reads from 454 sequencing are represented in a completely different manner. That topic will be covered in a different commentary.