
Six Tutorials in Various Stages of Completeness
We decided to post all tutorials in their current forms in our tutorials section and continue to work on them to polish up. Some of them should not posted at all, because they contain plans for writing rather that actual text. Let us tell you at the outset what we are trying to do so that our readers and we remain on the same page.
About two years back, we started writing on bioinformatics in this blog after realizing that bioinformaticians were focusing too much on downloading and running codes and less on actual algorithms. For example, if you were stuck with installing and running Velvet at that time, few websites, mailing lists and forums could help you out. However, when we searched for information on various steps of the algorithm, Zerbino’s thesis+paper and Velvet source code were the only places to go. We worked through the code and wrote few commentaries on de Bruijn graphs -
etc.
Our initial reason behind learning the algorithms was to speed up the execution and memory-handling of assemblers, because very few low memory assemblers existed at that time. Soon we discovered another advantage of knowing the algorithm in detail. Sometimes the assemblers are flat-out wrong, and genome assembly appears as black art, if the user does not understand what is going on. The same is true for nucleotide search, alignment and every other class of bioinformatics program. At that point, we decided not to try any bioinformatics algorithm without understanding its code or at least the core algorithm. We followed the same principle ever since, and more recently in trying the SGA assembler with great help from Jared Simpson.
The purpose of writing these commentaries has been to help out other bioinformaticians, who are possibly in the same boat as us. For example, it took us over two days to figure out all the ‘Algorithm’ blocks in the Fermi paper. If our writing helps you save even a day, that is great for open science. In general, the efforts have been appreciated by open science enthusiast like Titus Brown, who mentioned our blog at the top of the list in their recent MSU course on next-gen algorithms. Yeah right !!
Why the Tutorials?
After writing several commentaries on topics ranging from genome assembly to hardware and domestic spying, we started to feel the ‘discovery’ problem. Personally, we use the commentaries as our own notes on various topics, and go back to them from time to time to remember what we figured out during earlier round of thinking. Now that we have a large number of articles, we use google to get to our own writing, and we never seem to find what we are looking for. That is not only a problem with our own writing. Few minutes back, we searched google with keywords ‘titus brown rayan chikhi summer course’ to find a link the the summer course, and here is what we got.
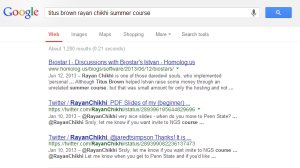
To address the issue, we decided to write few ‘index’ commentaries from time to time linking to various earlier commentaries. That created a different google problem. Whenever we searched for any topic in our blog, google always gave us the ‘index’ commentaries.
We realized that if we could not find our own commentaries, others would have even more difficult time finding what is written here. That led us to build the tutorial section. Right now the content has been split into six overlapping categories -
1. De Bruijn Graphs for NGS Assembly
This tutorial includes our earlier commentaries on de Bruijn graphs. A member of seqanswers described it as -
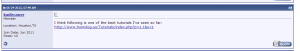
but we think our text can be improved substantially.
2. Algorithms for PacBio Reads
This tutorial focuses on all algorithms related to PacBio libraries. PacBio is being handled separately, because those indel-heavy reads need separate type of algorithms.
The quality is not good and we received few public and private criticisms. We will be addressing them as soon as possible.
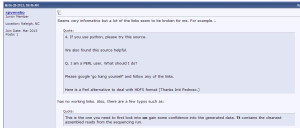
3. Software and Hardware Concepts for Bioinformatics
These tutorials will contain wide range of topics including hash function, Bloom filters, search algorithms, Hadoop. Also, all of our CPU, GPU and FPGA- related discussions will go there.
4. Finding us in Homolog.us (Search Algorithms)
This set of tutorials will focus on Burrow Wheeler transform and implementation of various NGS search algorithms. Few introductory commentaries we wrote in our earlier days are included here. As we work through BWA, SOAP3-dp and Bowtie2 algorithms, more texts will be added to these tutorials.
5. NGS Genome and RNAseq Assembly - a Hands on Primer
Knowing about de Bruijn graphs is one thing. Fully assembling a genome or transcriptome is another. We like to cover topics related to k-mer counting, error correction, assembly algorithm, scaffolding and post-processing here.
6. Introduction to PERL, Python, R and C/C++ for Bioinformatics
Should PERL go away through graceful death or should we stop language wars and start focusing on algorithms? Hopefully, we will be able to say something new using our ~30 years of experience in programming using every imaginable language, including byte-code, assembly and Verilog. This set of tutorials is the least complete.
-——————————————————–
A large part of the text is incomplete and highly unedited. We are slowly incorporating our previous blog posts into them, and we also plan to incorporate discussions of new blog posts there. We figured that keeping them posted in their current form and incorporating texts would be more helpful to the community than waiting for THE final polished form, which may take a while to happen.
Please tread carefully, while reading them. You have been warned :)
-———————————————————-
Why not Use Wiki and Crowd-sourcing?
We also built a wiki section, which is being used to explore large codes as in SOAPdenovo2. Wiki is a great tool for such tasks, but we do not enjoy wiki and especially crowd- sourcing for writing. Our ‘animal farm’ story will most be edited out as redundant in the first cut of crowd-sourced writing.