
Rosalind Project at Algorithmic Biology Laboratory, St. Petersburg
Nikolay Vyahhi, the author of rectangle graphs
paper,
emailed us about another project from their lab that our readers will find
very interesting. It is related to teaching bioinformatics
algorithmic biology.
You may be interested in another bioinformatics project from our lab called Rosalind, but it’s about an open education. It’s a free platform for learning bioinformatics through problem solving.
We announced it only in Russia in late August (just before RECOMB-BE 2012) and have about 400 active users learning bioinformatics at this moment. We plan to gradually scale it up to USA and world in a few months (just before winter quarter).
It also has a nice feature for bioinformatics professors to create a custom class using a subset of Rosalind problems, with automatic checking. TA’ing becomes an easy task. Though for the time being this feature is a bit hidden on profile page while we actively improve and polish it.
The website has a list of bioinformatics problems in the form of small puzzles. You can click on any problem, read its description, write a small script to solve it and enter the result. The website will tell you whether you are correct, and where you rank compared to others, who tried. Here is an example (happens to be among the most difficult)-
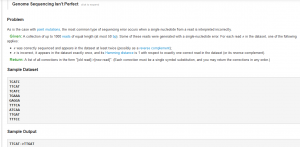
Please follow this link to try the problem.
We do not know, whether Dr. Pavel Pevzner has an active role in the above project, but there is good chance that the name ‘algorithmic biology’ came from him given how much he stresses on algorithmic thinking to solve biological problems (check Bob and Alice’s game). The aspect about algorithmic thinking is often missed in the typical bioinformatics analysis pipelines being used today. The end-user (senior biologist) outsources his Illumina sequencing and preliminary assembly to a core facility, and wants to receives greatly reduced data sets focusing on certain things he is familiar with. The core facility, on the other hand, tries to run many similar computations in a factory mode and does not have the capacity to explore unusual aspects of the data. As a result, the research project discovers at most the ‘known unknowns’, but rarely ventures into ‘unknown unknowns’ of the data.
Based on his writing, Dr. Pevzner does not want bioinformatics to be defined only as the ability to run BLAST, CLUSTAL, Bowtie, Velvet, etc. but rather to understand the algorithms behind those programs and their applicability in the biological context. Hopefully, the Rosalind puzzles will help students properly orient their thinking to that mode.