Various Developments - 11/30/2012
1. Can a Jellyfish Unlock the Secret of Immortality?
Well written article from NY Times, not a science journal. Next time I will ask a journalist to write my papers :)
[Note for would be critics - ‘well written’ is not equal to ‘scientifically correct’]
2. New Genomes are here - Watermelon and Wheat.
Watermelon paper is locked up by Nature Genetics, but you can see the supplem ent, which is possibly as informative than the paper.
Also check -
Watermelon Genome Decoded: Scientists Find Clues to Disease Resistant Watermelons
To download or visualize genomic data, please follow these two links - here and here.
Paper for wheat genome (17 Gb, hexaploid, 96000 genes, 80% of genome has repeats !!!) is accessible at Nature.
Which interesting plant genome remains to be decoded? Here is a good suggestion from an old (2010) blog post by James and the Giant Corn blog said
Pineapples use CAM photosynthesis. Normally plants have to open tiny holes in their leaves (called stomata) during the day to let in carbon dioxide that they use during photosynthesis. The problem they face is that when theyre letting carbon-dioxide in, plants also let water out.
CAM plants get around this water loss by collecting all their carbon dioxide at night (when its not as hot so they lose less water when they open their stomata) and storing it within their leaves until they need it during the day. This allows them to be much more efficient with water than normal plants (ones carry out plain old vanilla C3 photosynthesis.*)
….
With all these new third generation sequencing technologies coming out in 2010, hopefully someone will sequence the pineapple genome.
3. Oculus: faster sequence alignment by streaming read compression
Here we present Oculus, a software package that attaches to standard aligners and exploits read redundancy by performing streaming compression, alignment, and decompression of input sequences. This nearly lossless process (>0 99.9%) led to alignment speedups of up to 270% across a variety of data sets, while requiring a modest amount of memory. We expect that streaming read compressors such as Oculus could become a standard addition to existing RNA- Seq and ChIP-Seq alignment pipelines, and potentially other applications in the future as throughput increases.
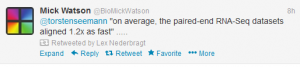
4. Oh no, yet another iProduct !!!
iReckon: Simultaneous isoform discovery and abundance estimation from RNA-seq data h/t @genetics_blog
In this paper, we introduce iReckon, a method for simultaneous determination of the isoforms and estimation of their abundances. Our probabilistic approach incorporates multiple biological and technical phenom- ena, including novel isoforms, intron retention, unspliced pre-mRNA, PCR amplification biases, and multi-mapped reads. iReckon utilizes regularized Expectation-Maximization to accurately estimate the abundances of known and novel isoforms. Our results on simulated and real data demonstrate a supe- rior ability to discover novel isoforms with a significantly reduced number of false positive predictions, and our abundance accuracy prediction outmatches that of other state-of-the- art tools. Furthermore we have applied iReckon to two cancer transcriptome datasets, a triple negative breast cancer patient sample and the MCF7 breast cancer cell line, and show that iReckon is able to reconstruct the complex splicing changes that were not previously identified.
5. Another Google Special that bioinformaticians would like -
Announcing Google Drive Site Publishing
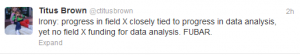
6. Humor - Law’s Laws for Bioinformatics h/t: @Assemblathon
This one is funny too except that it closely reflects reality -