
Study History and Read Papers Written by 'Dinosaurs' (#GWAS)

-————————————
Huge amount of money gets spent on genome wide association studies (#GWAS) with the promises that they will lead to finding cures of complex diseases, but how good are those promises? In today’s twitter brawl, BioMickWatson said that #GWAS contributed to human knowledge. Dan Graur wanted to find out, but he could not understand even the layman’s summary of a recent #GWAS paper. The following two entertaining posts present his experience:
#ENCODE, #GWAS, and #Pentecostalism: A true religion or a Sokal-like hoax
#ENCODE, #GWAS, and #Pentecostalism: Part 2
The author of the paper agreed that the summary was too complex and we offered a way out -
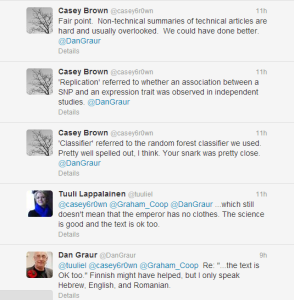
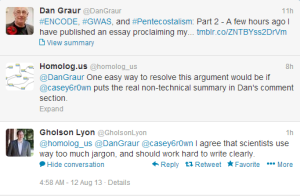
-—————————————–
What is the bigger issue here? Is Dan Graur complaining because he does not understand all these new cool developments, or is #GWAS another faddish branch of science as Ken Weiss argued? BioMickWatson tried to discredit Dan Graur’s opinion as ‘out dated dinosaur’, but we believe that is very disingenuous.
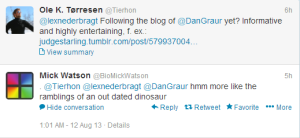
Dan Graur continues to do active research in population genetics and evolutionary biology, and was an author of the paper on genome comparison of social insects that we linked the other day.
Is Social Behavior Hardware or Software Coded?
So a better way to discredit him would be to say that evolution and population genetics are irrelevant for #ENCODE and #GWAS, which is Dan’s point as well (#ENCODE is closer to ID than evolution). On the relevance of population biology to #GWAS, Dan Graur presented one paper, and even its abstract shows (emphasized by us) where the conflict between the ‘cool new things’ and ‘dinosaur’s research’ lies.
Population genetics is fundamental to our understanding of evolution, and mutations are essential raw materials for evolution. In this introduction to more detailed papers that follow, we aim to provide an oversight of the ?eld. We review current knowledge on mutation rates and their harmful and bene?cial effects on ?tness and then consider theories that predict the fate of individual mutations or the consequences of mutation accumulation for quantitative traits. Many advances in the past built on models that treat the evolution of mutations at each DNA site independently, neglecting linkage of sites on chromosomes and interactions of effects between sites (epistasis). We review work that addresses these limitations, to predict how mutations interfere with each other. An understanding of the population genetics of mutations of individual loci and of traits affected by many loci helps in addressing many fundamental and applied questions: for example, how do organisms adapt to changing environments, how did sex evolve, which DNA sequences are medically important, why do we age, which genetic processes can generate new species or drive endangered species to extinction, and how should policy on levels of potentially harmful mutagens introduced into the environment by humans be determined?
It is possible that those ‘old’ notions of population genetics are wrong, but in science, the new guys have the responsibility to actually prove them wrong before making bold new statements.