
Reporting on the Northwest Developmental Biology Conference - (2)
New readers can follow us on twitter - @homolog_us
Our previous commentary was on number of general observations regarding NWDB conference. In this commentary, we cover a tiny subset of outstanding talks and presentations. The sampling is completely arbitrary and not representative of anything apart from my own ignorance. The presentations covered topics in biology, but since our readers are mostly from bioinformatics, I also speculate on how a bioinformatician can contribute to each topic, and you will find those discussions related to the final paragraph of the previous commentary.
I also sent requests to the speakers for their slides and three of them sent theirs. I will add the remaining ones, if I get them, but please note that the culture in biology is a bit different from what we bioinformaticians are used to.
-——————————————–
1. Maria Barna
In “Evomics Conference and a Paper by Carl Woese”, we mentioned a 2004 paper titled “A New Biology for a New Century” by Carl Woese. In that paper, Woese predicted that our new century would finally see ribosome as a regulatory system and not merely as an ‘inert’ translation machine. He made his statement based on the observation that the complex translation machinery had to appear from somewhere through a long evolutionary process, and the vestiges of those pieces must remain in the contemporary cell. Given that the RNA world had its own (RNA) enzymes and regulatory system, it is likely that its efficient components are still present in cells and undiscovered.
It was fascinating to find Maria Barna living that ‘ribosome is a ribozyme’ reality, and apply the concepts to one of the most important classes of developmental genes - Hox cluster. To learn about her work, readers can start with the following paper, but she shared a number of unpublished results in her talk. Overall message - protein translation done by ribosome is regulated by specific sequence in UTR or other coding regions. Mess up with those and you can end up with developmental defects.
Ribosome-Mediated Speci?city in Hox mRNA Translation and Vertebrate Tissue Pa tterning
Historically, the ribosome has been viewed as a complex ribozyme with constitutive rather than regulatory capacity in mRNA translation. Here we identify mutations of the Ribosomal Protein L38 (Rpl38) gene in mice exhibiting surprising tissuespecific patterning defects, including pronounced homeotic transformations of the axial skeleton. In Rpl38 mutant embryos, global protein synthesis is unchanged; however the translation of a select subset of Homeobox mRNAs is perturbed. Our data reveal that RPL38 facilitates 80S complex formation on these mRNAs as a regulatory component of the ribosome to confer transcript-specific translational control. We further show that Rpl38 expression is markedly enriched in regions of the embryo where loss-of- function phenotypes occur. Unexpectedly, a ribosomal protein (RP) expression screen reveals dynamic regulation of individual RPs within the vertebrate embryo. Collectively, these findings suggest that RP activity may be highly regulated to impart a new layer of specificity in the control of gene expression and mammalian development.
What can a bioinformatician do here?
Barna showed most of her data in mouse and mentioned that the regulatory sequences were NOT conserved over any meaningful evolutionary distance. That was quite puzzling, because it is hard to believe that similar patterns are not present in other organisms. Maybe sequence conservation is not the way to go and the nucleotides maintain different form of conservation (structure?).
Thinking about the other end of the spectrum, in yeast we found a very strange RNA several years back. it was hidden in antisense region of a protein-coding gene, but was functionally different from the protein-coding gene. Moreover, Sid Altman, who received Nobel prize for discovering ribozyme RnaseP, followed up on our work and showed that HRA1 was a substrate of Rnase P. Rnase P processes precursor tRNAs, which Woese expected to turn out to be important block of his regulatory ribozome world. No further work has been done on HRA1.
By looking at those various pieces, it is clear to me that the RNA world is far from understood in many different organisms and a lot remains to be discovered. Bioinformaticians have an open field to explore in figuring out how evolution works in these contexts !!
-——————————————–
2. Elizabeth Haswell

Moving to one level higher to proteins, Elizabeth Haswell presented on mechanosensor proteins on the E. coli cell membrane and plant cell walls. Her work nicely combines biology and biochemistry (structure). Borrowing from her website -
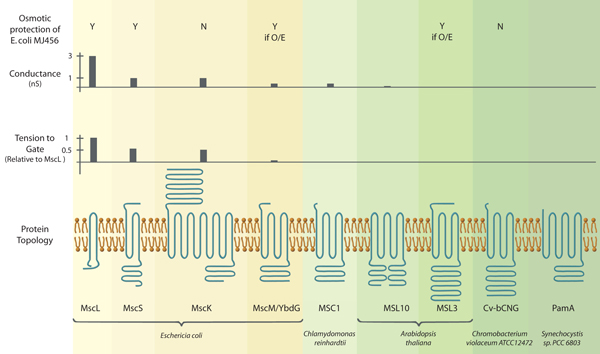
Photo caption: (Functional and structural characteristics of MscL and MscS channels from bacteria and eukaryotes. (Top panel) Expression of MscS and MscL protects a bacterial strain lacking endogenous MscS and MscL from a 0.5 M osmotic downshock while expression of MscK and Cv-bCNG does not (Levina et al., 1999; Caldwell et al., 2010). Asterisks, over-expression of YbdG and MSL3 provides protection (Haswell et al., 2008; Schumann et al., 2010). (Top middle panel) Conductance of endogenous channels in giant E. coli spheroplasts (MscL, MscS, MscK, MscM, (reviewed in (Kung et al., 2010))), channels heterologously expressed in giant E. coli spheroplasts (MSC1, (Nakayama et al., 2007)) or endogenous channels in Arabidopsis thaliana root cells (MSL10, (Haswell et al., 2008)). (Bottom middle panel) Tension to gate expressed relative to MscL channels in the same patch (Berrier et al., 1996; Edwards et al., 2005; Li et al., 2007). (Bottom panel) Channel monomer topologies as predicted by TOPCONS (http://topcons.net/). Mature versions (after processing of chloroplast targeting sequences) of MSC1 and MSL3 are shown, and sequence loops connecting transmembrane helices were omitted for clarity.)
Haswell kindly shared a subset of her slides with our readers -

What can a bioinformatician do here?
Bioinformaticians often analyze gene expression and perform GO analysis, but her work showed what needed to be done to properly understand each of those over/under-expressed protein families. Possibly a bioinformatician can follow through all details provided by her and try to replicate the evolutionary analysis part in other gene families. For a good example, readers are advised to follow the work from Yves van der Peer’s group as shown below.
Haswell’s discussion of bacteria and plants reminded me of Thomas Cavalier Smith’s paper “Eukaryote kingdoms: Seven or nine?” and I got curious about what happens in Chromophyta. The relevant data were available from a genome project that I took part in several years back, and I fould what I was looking for in the supplement, thanks to Yves van der Peer’s group.
The Ectocarpus genome and the independent evolution of multicellularity in brown algae
Three genes encoding bacterial-type, small conductance mechanosensitive (MscS) channel are present in the Ectocarpus genome. These channels are involved in osmoregulation in E. coli310, chloroplast development in Chlamydomonas and Arabidopsis311,312, and root hair mechanosensation in Arabidopsis313, and could be involved in environmental sensing in brown algae, particularly in relation to osmosensing in the intertidal environment314. The tandemly repeated genes Esi0032_0081 and Esi0032_00822 are most similar to an Arabidopsis MscS domain containing protein, Y5208_ARATH. The third Ectocarpus MscS channel is most similar to the yeast MscS yna1 channel. A survey of the sequenced stramenopile genomes indicates that the diatom genomes contain more MscS channels than Ectocarpus (Supplementary Table 38), arguing against a role for these channels in the evolution of multicellularity. No sequences representing the large conductance mechanosensitive channels were identified in the Ectocarpus genome.
-——————————————–
3. Norbert Perrimon
Moving from individual protein families of Haswell to protein-protein interactions, Norbert Perrimon from Harvard invented the GAL4 reporter many years back and revolutionized the fly world. The technique invented by him is described as fly geneticist’s Swiss army knife (GAL4 System in Drosophila: A Fly Geneticists Swiss Army Knife).
That is only one of his contributions. In the NWDB conference -
(i) He presented unpublished work on a very cool dynamic reporter system based on combining stable RFP and unstable (superfast) GFP. This allowed him to measure time-course pattern of wnt expression at a very fine timescale.
(ii) He also presented on using his various reporter systems to understand the inter-organ communication in fly. Those results are already published.
**How can a bioinformatician contribute? **
Apparently plenty, because his lab also developed a large library of Drosophila RNAi and is trying to functionally annotate all protein-coding genes and find protein clusters. Several years back (2003), we did some bioinformatics work on similar topic, when the yeast community was at the same stage.
Predicting protein functions from redundancies in large-scale protein interaction networks
One surprising finding at that time was that most of the unannotated proteins were involved in translation and RNA-processing pathways. Norbert mentioned that he did not see any such pattern in his newly derived interactions in fly. It is either we learned a lot more about proteins (but not RNA) in translationary machinery by now, or the functional annotations derived by us propagated to other databases.
-——————————————–
4. Tamily Weissman

Going one step higher from cells and cellular signaling, Tamily Weissman is famous for her work on brainbows for measureing neuronal patterning. In her talk, she described the implementation in zebrafish.
Brainbow is the process by which individual neurons in the brain can be distinguished from neighboring neurons using fluorescent proteins. By randomly expressing different ratios of red, green, and blue derivatives of green fluorescent protein in individual neurons, it is possible to flag each neuron with a distinctive color. This process has been a major contribution to the field of connectomics, or the study of neural connections in the brain. The study of neural pathways is also known as Hodology by earlier neuroanatomists.
**How can a bioinformatician contribute? **
At this point, they look into the growth of neurons through bare eyes, but that could be tedious for large and complex brains. It could be possible to use signal processing algorithms of electrical engineering to develop an automated approach.
-——————————————–
5. Eric Haag


Eric Haag’s work goes right to the heart of natural selection. He had been looking into genome sizes of several members of genus Caenorhabditis (worm) and found that the genomes of hermaphrodite worms go through early deletion. More details (including slides) on his talk are given in a separate blog post, but here is a brief description of his research from his webpage.
My laboratory studies the developmental genetics of evolutionary change in animals. Of particular interest to me are reproductive adaptations whose evolution required major developmental novelties. Currently the lab is focused on the evolution of self-fertile hermaphroditism in nematodes. We use the model nematode Caenorhabditis elegans as a starting point, which confers several advantages. First, sister species of C. elegans have different reproductive modes that are based on differences in sex determination in a single tissue–the germ line. Second, C. elegans sex determination has been subjected to intense genetic, molecular, and biochemical investigation, which provides a wealth of potential mechanisms for investigation. Third, many of the tools available to C. elegans researchers are applicable to its relatives as well, such as classical genetics, RNA interference-mediated reverse genetics, and even complete genome sequence.
We seek to identify the molecular and genetic mechanisms that distinguish the sex determination of androdioecious (hermaphrodite/male) species from that of gonochoristic (male/female) species of worms. We also are interested in the convergent evolution of selfing, the evolutionary forces that drive the rapid evolution of sex determination even in the absence of overt phenotypic change, and the molecular and genomic responses to these forces.
What can a bioinformatician do here?
The sex-related parts of most genomes are not well-annotated and often not even well-assembled, even though that is where the most interesting information about natural selection possibly lie. Moreover, there is tremendous diversity and almost nobody looked at the sex chromosomes from the genome evolution angle. I think bioinformaticians can contribute tremendously here. Another question comes to my mind - will it be possible to combine the ‘RNA world’ with diversity of sex-related genes? Plenty remains to be uncovered.
-——————————————–
6. Scott Freeman

Scott Freeman was invited to present on a somewhat different topic - ‘active learning’. The message of his talk was that straightforward lecturing is inefficient, and a Socratic method of learning works much better. The method derived by him is a combination of Socratic method and modern electronic technology. Moreover, when he says ‘works much better’, he backs those statements with plenty of data and analysis.
Freeman very generously shared his slides -

What can a bioinformatician do here?
I asked him whether MOOCs are any good and he said possibly not, as long as they are lecture-based. Later, I have been wondering whether his method is applicable to MOOCs and realized that Rosalind is indeed an implementation of his method of ‘active learning’. Then I also remembered that another method for learning bioinformatics called Socratique also exists. Possibly bioinformaticians are already into this game of innovation. If you are not familiar with Rosalind or Socratique, please check the following posts -
Rosalind Project at Algorithmic Biology Laboratory, St. Petersburg
Rosalind Will Get Even More Marvelous
On Teaching/Learning Bioinformatics Online Socraticqs, Rosalind
