
Watching a de Bruijn Assembler in Action (Contrail)
_Job opening at Bermuda Institute of Ocean Sciences- analysis of microarray data
A collaborator of us from the sea urchin community emailed about a job opening in her group. You can find the details about the position here. If interested, please feel free to contact her at Andrea.Bodnar at bios.edu.
_
-—————————-
We wrote many commentaries on how de Bruijn graph based assemblers work - forming the initial graph, correcting for errors that form hanging chains or ‘bubbles’, assembling around repetitive regions and building larger scaffolds using paired end reads. Wouldn’t it be great to see these steps in action? In our commentaries, we can only work out one or two examples in detail, but it would be far more beneficial to start from your favorite set of reads and see how a de Bruijn assembler works through them to give you the assembly.
Unfortunately, most assemblers do these calculations in memory and give you the final result or intermediate results only at important junctures. That is understandable, because writing a large graph on the disk costs time. Contrail is the only exception in this regard. Contrail uses disk-based Hadoop, and not huge RAM, to assemble a genome. Therefore, it operates by writing an image of the graph at every step and then letting the next stage of map-reduce to take the graph as input and process it. This provides great opportunity for anyone to play with small examples and learn about various steps of the algorithm.
The following figure shows important directories, if you want to understand Contrail output. I left out few other directories that I believe are more related to giving stats about the graph than storing the graph itself. Once again, we are not the authors of this program or associated in any way to their group. We are figuring out the details by reading the code and outputs, and may miss a step or two.
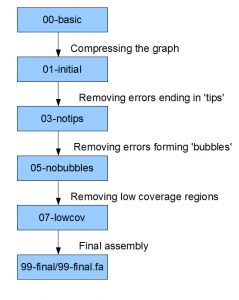
As we mentioned previously, Contrail can be run by setting up Hadoop on a single machine. In fact, we installed hadoop installed in a windows PC under cygwin, and often run Contrail there to check how it works on small test libraries of reads.
When you see the Contrail output graph for the first time, the characters may appear confusing. You will see many Ds, Ms, Qs, Fs and other characters from A-T instead of our favorite A, T, G, C. There is nothing scary; Contrail replaces nucleotide pairs into letters from A-T to save space. We wrote a perl code to go from A-T space to A,T,G,C that we shall upload here shortly.