
Modularizing SOAPdenovo2 Code into Four Components
We discussed the code of SOAPdenovo assembler in January and ended with -
Here is our bigger objective. We plan to split all major assemblers into code components and mix and match them to assemble the fish library in Assemblathon 2, which was the hardest to assemble. We like to understand the results of Assemblathon 2 at the algorithmic level, and explain to our readers.
In between other work kept us busy, but finally we managed to go through the SOAPdenovo2 code and split it into four components.
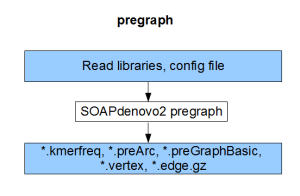
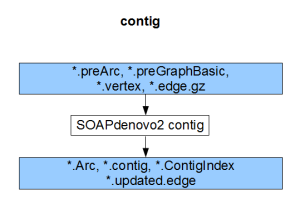
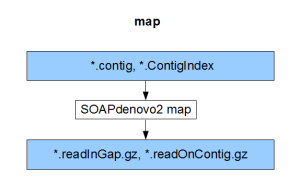
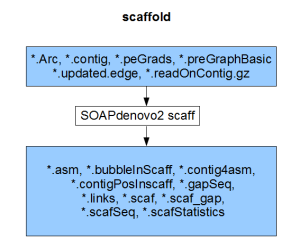
Each component compiles on its own and gives the result of the respective stage (pregraph, contig, map, scaff). So, if we can generate identical set of outputs from a different assembler, those files can be fed into the remaining stages of SOAPdenovo2 to complete the assembly.
The code of SOAPdenovo2 uses pthreads, not openMP. That allows finer level of control over the parallel blocks and thus faster execution. MPI codes, on the other hand, are easier to understand and update.
What can we do with the modules? Here are the possibilities -
i) Replace pregraph stage with Minia-like Bloom filter algorithm to reduce memory (RAM) requirement,
ii) Try multi-kmer approach, which does not work well with the current SOAPdenovo2. There is no guarantee that we can do better, but we have some ideas.
iii) Try rectangular graph style algorithm along with the current scaffolder.
iv) Fool the scaffolding unit with PacBio reads to try hybrid assembly.
v) If everything else fails, we can at least make the modified assembler print a wisecrack every few minutes, while you are waiting for the results. That will be The Onion version of a genome assembler :)
We are trying to figure out a way to post the above code in github without creating too much confusion with the original version.