
Keith Robison's Perfect Scaffolder Delivered, What Next?
On January 1st, Omicsomics blog requested for the perfect scaffolder.
Envisioning The Perfect Scaffolder
Rather than make any New Year’s resolutions of my own, which I would then feel guilty about not keeping, I’ve decided to make one for someone else: they will write the perfect open source scaffolder. There’s a lot of scaffolders out there, both stand-alone and integrated into various assemblers, but none are quite right.
On March 6th, a benchmarking paper from Sanger Institute delivers it, provided…
A comprehensive evaluation of assembly scaffolding tools
We further dissect the performance of the scaffolders using real and simulated sequencing data derived from the genomes of Staphylococcus aureus, Rhodobacter sphaeroides, Plasmodium falciparum and Homo sapiens. The results from simulated data are of high quality, with **several of the tools producing perfect output.
….
Overall, SGA, SOPRA and SSPACE generally outperform the other tools on our
datasets.**
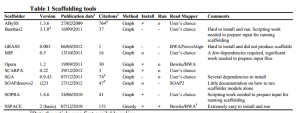
The paper seems to have done a very thorough job by checking each scaffolder based on multiple aligners. The choice of aligner matters !!
P. S.
1. Please note that the subject line is written in jest. Real scaffolders on real data perform quite lousily given that ‘90% correct’ scaffolding is still a bad situation for those 10% of erroneous parts of the genome.
2. Wish the paper could compare SPAdes along with other scaffolding methods.