
Oxford Nanopore Fans are Asking Three Questions Today
1. Price
The company announced the prices for their sequencers and many people are not happy.
Oxford Nanopore said last month that it plans to make the MinIon sequencer commercially available through the MinIon Access Program. Customers need to join the MAP, which the firm describes as a developer-style access program, and pay a $1,000 fee, which includes the MinIon MkI device and a starter kit with three flow cells, two reagent kits, software, and ongoing intermittent free supplies.
Users can purchase additional flow cells in sets of 1, 12, 24, or 48, at a cost ranging from $500 per flow cell to $900 per flow cell, depending on the number of flow cells purchased at a time.
Torsten Seeman noted -
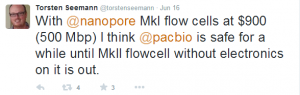
Mike the Mad Biologist noted -
A Question About Oxford Nanopore and the Cost Model
If we limit ourselves to best case scenario of what is actually in hand, 1Gb of sequence at $500, that doesnt seem so hot. In the microbial world (cuz humans are boring), that might get you two or three bacterial genomes per flow cell, and that would then require some sort of barcoding method (MOAR MONIES!) to tell samples apart. Otherwise, you are, in the best case, spending $500 on sequencing reagents alone per bacterial genome. You might as well do PacBio sequencing.
That is quite a bit of change in sentiment from last year, when one dyed-in- the-wool nanopore fan privately advised Jason Chin (bioinformatics director of Pacbio) to look for another job.
-——————————————
2. Quality
What is the real error rate of Oxford Nanopore reads? Is 500Mb of nanopore data equivalent to 500Mb of Pacbio data? All published literature we have seen so far argued that Oxford nanopore reads were inferior by several notches. Ask yourself this. If the error rates were identical, why would nanopore analysis need a more sensitive aligner (LAST) and an elaborate assembly procedure?
Mike the mad biologist seems to have the same complaint -
Im not convinced at all that for routine surveillance (not whaddya think about these six isolates) the error rate for Nanopore allows you to identify incipient outbreaks. Too many errors as best as I can tell.
-——————————————-
3. Perpetual MAP with no criticism?
Let us take a step back and look at the history of this company. In 2014, the company kicked out Alex Mikheyev from its ‘early access’ program, because his paper talked about poor read quality. The company claimed that no criticism would be allowed in the MAP phase.
Open Science Fail Oxford Nanopore Kicks Out Alex Mikheyev from its MAP Program
Our reader David Eccles has been providing steady updates about his progress with the technology (see all comments in Nanopore Updates from David Eccles), and here is what he wrote after the London Culling meeting.
One of the false myths that Ive been spreading round is that the MinION was not yet commercialised. In fact, Gordon Sanghera said at the opening that MAP would never end, and I had confirmation from Roger Pettet that MAP is effectively the commercialisation of the MinION.
That is in agreement with the latest announcement. MAP program is perpetual.
Oxford Nanopore said last month that it plans to make the MinIon sequencer commercially available through the MinIon Access Program. Customers need to join the MAP, which the firm describes as a developer-style access program,
Does that mean the ‘no criticism’ policy is also perpetual? Is Mike the Mad Biologist out for not following the party line? Users would like to know.
-———————————————————
Edit. Lex Nederbragt, who created a beautiful figure to compare sequencing technologies, added a fourth question in his blog post.
Developments in high throughput sequencing June 2015 edition
The Oxford Nanopore MinION is a bit tricky. My metrics are based on company specifications that anyone can view from their website, for commercially released instruments and (chemistry) updates. The commercial release of the MinION seems now to have happened, as it was announced everyone can apply for the MinIon Access Program and will be accepted (barring some sanity screening they probably will do). But metrics for this instrument are a different matter. There are no company specs that I can find. Partly this is understandable, as read length, for example, is dependent on the input length of the sample (or library, rather). The other reason for the lack of specifications may be the Minion Access Programs philosophy of Oxford Nanopore. It is the users that are discovering what the instrument can do, rather than the company telling the customer what to expect.
-—————————————–
Edit.
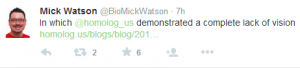
BioMickWatson, a ‘visionary’ nanopore promoter, fails to answer any of the questions - **(i) Does perpetual MAP mean perpetual ‘no criticism’ policy? (ii) What is the current read error rate? (iii) When exactly should I throw away the Pacbio machine? **
He responded with -
“Wondering how you can fit MinION into your existing workflows is like sitting in front of a space ship and wondering how youre going to use it to commute to work”.
Geez !! Someone like me, who worked for NASA for six years, knows very well that the cost and the quality of ride matter as much as having a ‘space ship’. In 2004, Bush unveiled ‘moon to mars’ mission. However, internally at NASA, we came to the conclusion that such a thing would be impossible, because it appeared too damn expensive. Moon to mars was canceled without fanfare after a few years.
Therefore, I would replace the cute quote by Biomickwatson with another more appropriate one -
When a man with money meets a man with vision, the man with vision leaves with money and the man with money leaves with vision.