
Pacbio Assembly - Miniasm Works Best (Chaisson)
A few months, we wrote about Heng Li’s Minimap and Miniasm. Yesterday, Mark Chaisson posted a chart in twitter showing that they perform really well for assembling pacbio reads. Miniasm is excellent even with low coverage.
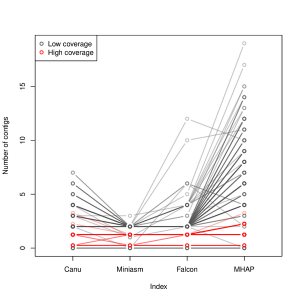
-———————————-
Edit.
Various caveats from Chaisson and Jason Chin -
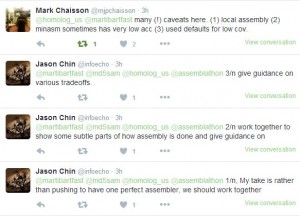
-—————————————
Martin Hunt came to a different conclusion, as pointed out by @md5sam in twitter.
It is neat that we now have a tool that can completely skip read error correction and still produce not just any output, but good output. miniasm is absolutely worth running and the quality of its output came as a pleasant surprise, especially considering that the reads are not corrected and it has short run times and low RAM usage. Even better, it did not make the false joins and large inversion errors that were made by HGAP. In general I prefer a conservative, more fragmented, assembly that has no errors. miniasm often made a better job of the plasmids, compared to HGAP. However, miniasm struggled on two samples, producing a large number of contigs compared to HGAP.
For eukaryotic genomes, where even running read correction can be a major headache due to extreme CPU and RAM requirements, miniasm should provide a computationally feasible method of getting a first-pass assembly. How well it handles larger genomes with more repeat content remains to be seen.
Finally, as expected, the miniasm output obviously needs polishing. I presume that quiver will do the job, but likely with a longer run time than would be needed on assemblies made from corrected reads. That is work for another day.
-———————————-
Further update from Martin Hunt -
Please check the github directory here.