
Explaining Output of Trinity Component - Inchworm
The transcriptome assembler Trinity from Broad Institute runs through three steps - Inchworm, Chrysalis and Butterfly. Inchworm is the first program of the pipeline. It operates on the RNAseq library to produce many thousands of sequence fragments. What is the relation between those sequence fragments and the actual genes?
We explained in our earlier commentary that, for an alternatively spliced gene, inchworm prints the entire length of highly expressed form as one sequence, whereas only parts of other splice forms are given as other sequences. It was written entirely based on our understanding of the steps of the algorithm. So, today we ran a simulation to check whether inchworm really behaves the way the algorithm suggests it would.
For this simulation, we created two 4KB genes by stringing together random nucleotides. Both genes have the first and the last 1500 nucleotides as identical, but the middle 1000 nucleotides as different.
In the first simulation, we picked 4000 75-mer short reads from the first gene (Gene 1) and 2000 75-mer reads from the second gene (Gene 2). Inchwork had two outputs - (i) the entire structure of Gene 1 except one nucleotide at the end and (ii) the middle 1KB (1047 nucleotides) of Gene 2. In terms of the following figure, one inchworm output consists of A+X+B from Gene 1, and the other inchworm output consists of only Y.
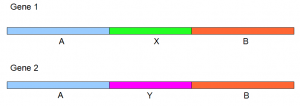
In the second simulation, 4000 reads were picked from Gene 2 and 2000 from Gene 1. Inchworm output reversed. We got (i) (i) the entire structure of Gene 2 except two nucleotides at the ends and (ii) the middle 1KB (1047 nucleotides) of Gene 1. In terms of the above figure, one inchworm output consists of A+Y+B from Gene 2, and the other inchworm output consists of only X.
What is the significance of 1047 number? Why did not inchworm give exact 1000 nucleotide long segment? The size 1047 comes from 1000 + 2* (K-mer length-1). Inchworm uses 25-mers in its default set up and so it includes all 25-mers that overlap with the middle section, but do not fully overlap with the other gene. If we increase the K-mer length to 31, inchworm would likely give 1059 or 1060 nucleotide long middle segment. This point is important in understanding the algorithm of other two steps of Trinity.