
Output of Oases Transcriptome Assembler
We discussed how to run Oases transcriptome assembler in an earlier post. In this exercise, we ran the same simulated example through both Velvet+Oases pipeline and Trinity pipeline to highlight their key difference. Inchworm results were discussed in the previous commentary, where we also discussed how the genes were constructed. Readers are encouraged to read the previous commentary before proceeding with this one.
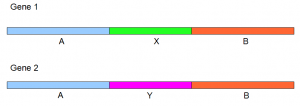
When the above gene was run through Velvet+Oases, Velvet produced four contigs
- A, B, X and Y. They were stored in Velvet output file ‘contigs.fa’. You see the major difference between Velvet+Oases and Trinity here. Inchworm, the first step of Trinity, gave the complete gene structure A+X+B and the additional fragment Y.
After the Oases program processed Velvet output, Oases correctly produced two alternative structures of the same gene. They were written in the file ‘transcripts.fa’.
Oases also created a file titled ‘contig-ordering.txt’, where it showed how the Oases transcripts and the Velvet contigs are related. It produced entries like -

This is the same gene structure that we showed in the figure above. It says that Transcript 1 is formed of contigs 3–>1–>2, and Transcript 2 is formed by 3–>4–>2. So, instead of A, X, B, Y in our notation, Oases named them contig 3, contig 1, contig B and contig Y.
The other Oases output file ‘splicing-events.txt’ presents the same information in a different format.