
CHANCE - A Comprehensive and Easy-to-use Software for ChIP-seq QC
Reader Aaron Diaz from Jun S Song’s lab in UCSF alerted us about their recently published paper on new software for quality control and validation of ChIP-seq data. We never wrote about ChIP-seq and thought this would be a good opportunity to mention few other key papers as well.
CHANCE Download
Description
Based on description in the paper, the program developed by Aaron stands out in two respects - (a) in its sheer number of new features compared to other programs, (b) in its user-friendly graphical interface to make analysis easy.
From their abstract:
CHANCE (CHip-seq ANalytics and Condence Estimation) is a standalone package for ChIP-seq quality control and protocol optimization. Our user-friendly graphical software quickly estimates the strength and quality of immunoprecipitations, identies biases, compares the user’s data with ENCODE’s large collection of published datasets, performs multi-sample normalization, checks against qPCR-validated control regions, and produces informative graphical reports.
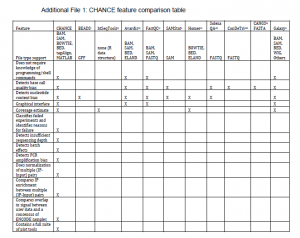
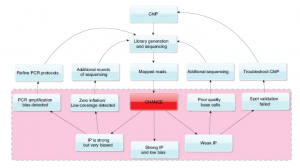
However, the best description of CHANCE’s features comes from supplementary CHANCE feature comparison table and Fig. 8 in the paper, as shown below.
Song lab at UCSF
We also checked the webpage of Song lab at UCSF and found it to be a collection of brilliant people, all from theoretical physics and mathematics background. Those interested in systems biology of transcription regulation may enjoy the following notes from Dr. Song’s lecture.
Key ChIP-seq Analysis Papers
While on the topic of ChIP-seq, we wanted to list few old and new papers useful for data analysis.
[Genome-Wide Mapping of in Vivo Protein-DNA Interactions, David S. Johnson, Ali Mortazavi, Richard M. Myers, Barbara Wold, Science 2007.
It was the earliest ChIP-seq paper. Following its links at google scholar is a good method to track the history of the field.
Model-based Analysis of ChIP-Seq (MACS), Yong Zhang et al. Genome Biology 2008.
It is among the most cited papers for analyzing ChIP-seq data. Shirley Liu’s group developed programs for analysis of microarray data, and this one extended the methods.
It is a good review paper we do not like to cite for its $32 cost. We wanted to move on to the next one, but thanks to the Chinese, found a free copy for you.
Absolutely fascinating paper. Authors not only compared all existing ChIP-seq analysis tools available in 2010, but had enough sense to publish their findings in PLOS One so that others can read.
Other ChIP-seq Analysis Papers
Choices of the following papers are arbitrary. They do not provide a full list of all relevant papers, but only a subset. Please feel free to add in comment section, if you think we missed a relevant and interesting paper that our readers may enjoy.
Our Paper
Finally, our modest contribution to the world of ChIP-chip and ChIP-seq:
The above research was done by the group of Angelike Stathopoulos, who is a very smart biologist at Caltech working on figuring out the transcriptional network in Drosophila.