
A Comparison of Two Turtle Papers

Green turtle

Painted Desert Turtle
In previous commentaries, we mentioned about two turtle genome papers published in this month.
i) Turtle the Luckiest Animal of the Month
ii) Genome of Western Painted Turtle.
Their journal links are here - here (Nature Genetics - NG) and here (Genome Biology - GB). The GB paper assembled the genome of western painted turtle using Sanger technology. The NG paper was actually a ‘buy one, get one free’ deal. It assembled the genomes of soft-shell turtle (Pelodiscus sinensis) and green sea turtle (Chelonia mydas). Anyone, who can guess the NGS-based one, will get 10 homolog.us points.
In this context, we can report an interesting experience we had with the authors of the GB paper. When the paper came out, we sent email to its first author asking some innocuous questions, such as when the project started, how much it cost to sequence the genome, what they plan to do to figure out the aging-related genes, etc. When we write about genome papers, we ask such questions to others, and as you remember, Dr. Amemiya told us about the trouble he had to go through to get coelacanth DNA sequenced. Dr. Shaffer was very friendly and tried to answer our questions, but higher authorities at The Genome Institute intervened and stopped all communication.
Few mundane stats -
Number of authors
GB=59, NG=34.
Submission date
GB:
Submission date 18 October 2012
Acceptance date 19 March 2013
Publication date 28 March 2013
NG:
Received 04 November 2012
Accepted 27 March 2013
Published online 28 April 2013
Genome size and N50
GB paper assembled 2.59 Gb genome with scaffold N50 around 5.2Mb covering at least 93% of the western painted turtle genome. NG paper reported both genomes of size around 2.2 Gb with the scaffold N50 >3.3 Mb.
Evolutionary origin of turtles
Apparently the evolutionary origin of turtles was not clear. Quoting the NG paper -
Three major hypotheses have been proposed for the evolutionary origin of turtles, including that they (i) constitute early-diverged reptiles, called anapsids, (ii) are a sister group of the lizard-snake-tuatara (Lepidosauria) clade or (iii) are closely related to a lineage that includes crocodilians and birds (Archosauria). Even using molecular approaches, inconsistency still remains.
Both papers did very nice comparative analysis of a number of vertebrates, and came to the same conclusion that turtles were closely related to crocodiles and birds.
Gene Family Comparison
This is where the papers started to diverge. We are not sure whether the divergence is due to difference in analysis, difference in genome assembly or difference in genomes.
NG paper reported a surprising finding.
Unexpectedly, we found that the olfactory receptor family was highly expanded in both turtle species. In particular, the soft-shell turtle contained 1,137 intact, possibly functional olfactory receptor genes, a number comparable to or even greater than the number of olfactory receptor genes found in most mammals.
It is also surprising that the GB paper did not report anything unusual about olfactory genes. Are those genes missing from the genome of western painted turtle?
Instead of discussing other differences, let us jump to the 100 million dollar question.
Why do turtles live so long?
NG paper speculated -
Finally, a possible connection to longevity in turtles was also found (Supplementary Table 24); the most accelerated gene in turtles, showing evidence of positive selection (with the rate of nonsynonymous nucleotide substitutions exceeding the rate of neutral mutations, dN/dS ratio > 1), was microsomal glutathione S-transferase 3 (Mgst3; dN/dS = 5.68), which is reported to function in antioxidative stress, and disrupting the homolog Mgst3-like in Drosophila melanogaster reduces lifespan.
GB paper identified a different set of genes.
The genomic basis of longevity in turtles: One of the defining features of turtles as a lineage is their extreme longevity (many species live 100 years or more), and we used the Western Painted Turtle genome to investigate this quintessential chelonian feature. Based on previous work implicating the shelterin complex encoding genes in exceptional longevity in the naked mole rat[42], we evaluated (by BLAST searches of all available turtle sequence data including unplaced scaffolds, see Materials and Methods, Aging and longevity, Additional file 1, Table S12) the status of the shelterin complex in the Western Painted Turtle genome. Even with this comprehensive search, we were unable to find orthologs for three of the five genes (POT1, TERF2IP, TEP1) in the Western Painted Turtle. Given that TEP1 is also absent in birds, this result strongly suggests that turtles (and their sister-group, the archosaurs) do not share this longevity mechanism with the naked mole rat.
Slow rate of evolution or fast?
GB paper reports -
Our phylogenetic analyses confirm that turtles are the sister group to living archosaurs, and demonstrate an extraordinarily slow rate of sequence evolution in the painted turtle.
NG paper reports -
Our results suggest that turtle evolution was accompanied by an unexpectedly conservative vertebrate phylotypic period, followed by turtle-specific repatterning of development to yield the novel structure of the shell.
Are those observations consistent?
Accelerated evolution
GB reports -
Among gene families that demonstrate exceptional expansions or show
signatures of strong natural selection, immune function and musculoskeletal patterning genes are consistently overrepresented.
NG reports -
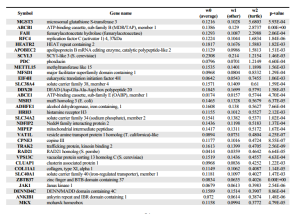

Given how much differences are seen between the genome analysis sections of two papers, we wonder whether it is meaningful to think about ‘the genome paper’ of any organism as the final verdict. How meaningful are the press releases either?
Painted Turtle Gets DNA Decoded
Turtle genome analysis sheds light on the development and evolution of turtle-specific body plan
We will continue to compare the papers and add other differences to this commentary.