
Is Seven Bridges Genomics Patenting All Existing Bioinformatics?
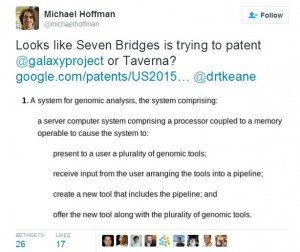
Bioinformaticians are up in arms after learning that Boston-based Seven Bridges Genomics and a sponsor of Titus Brown’s lab is trying to patent their published work. Zamin Iqbal pointed out in Twitter that the company applied for patent (in 2015) for graph-based alignment method that Weigel lab published in 2009. Michael Hoffman found out that Galaxyproject is also up for patenting.
Those twitter exchanges got Titus Brown defensive because of his financial ties with the company. Unlike his loud complaints about Kallisto’s non-free license (which Lior Pachter had every right to issue), this time his tweets seemed to be in support of the company.
In other news, leading scientist Tom Daschle joined the board of the company. Apparently, his connections with NIH will help the company win business. Daschle was nominated by Obama to be the HHS secretary in 2009, but had to quit after a ‘series of errors’ in his tax filing were discovered.
Daschle, the former Senate majority leader, apologized Monday for failing to pay his taxes in full. He said earlier he was “deeply embarrassed” for a series of errors that included failing to report $15,000 in charitable donations, unreported car service and more than $80,000 in unreported income from consulting.
For a full list of patent applications from Seven Bridges, you can check here.
Patent application number Title Published
20150227685 SYSTEMS AND METHODS FOR ANALYZING SEQUENCE DATA - The invention provides methods for comparing one set of genetic sequences to another without discarding any information within either set. A set of genetic sequences is represented using a directed acyclic graph (DAG) avoiding any unwarranted reduction to a linear data structure. The invention provides a way to align one sequence DAG to another to produce an alignment that can itself be stored as a DAG. DAG-to-DAG alignment is a natural choice wherever a set of genomic information consisting of more than one string needs to be compared to any non-linear reference. For example, a subpopulation DAG could be compared to a population DAG in order to compare the genetic features of that subpopulation to those of the population. 08-13-2015
20150112658 SYSTEMS AND METHODS FOR TRANSCRIPTOME ANALYSIS - The invention generally provides systems and methods for analysis of RNA-Seq reads in which an annotated reference is represented as a directed acyclic graph (DAG) or similar data structure. Features such as exons and introns from the reference provide nodes in the DAG and those features are linked as pairs in their canonical genomic order by edges. The DAG can scale to any size and can in fact be populated in the first instance by import from an extrinsic annotated reference. 04-23-2015
20150112602 SYSTEMS AND METHODS FOR USING PAIRED-END DATA IN DIRECTED ACYCLIC STRUCTURE - Methods of analyzing a transcriptome that involves obtaining at least one pair of paired-end reads from a transcriptome from an organism, finding an alignment with an optimal score between a first read of the pair and a node in a directed acyclic data structure (the data structure has nodes representing RNA sequences such as exons or transcripts and edges connecting pairs of nodes), identifying candidate paths that include the node connected to a downstream node by a path having a length substantially similar to an insert length of the pair of paired-end reads, and aligning the paired-end rends to the candidate paths to determine an optimal-scoring alignment. 04-23-2015
20150066383 COLLAPSIBLE MODULAR GENOMIC PIPELINE - The invention generally relates to tools for genomic analysis and particularly to a pipeline editor that can turn pipelines into standalone tools for use in other pipelines. The invention provides systems and methods for genomic analysis in which individual analytical tools can be arranged into analytical pipelines that can then be collapsed into standalone tools, which themselves can be put into the pool of individual tools for use in further building of pipelines. Aspects of the invention provide a system that includes a server computer system operable to present to a user a plurality of genomic tools, receive input from the user arranging the tools into a pipeline, create a new tool that includes the pipeline, and offer the new tool along with the plurality of genomic tools. 03-05-2015
20150057946 METHODS AND SYSTEMS FOR ALIGNING SEQUENCES - The invention includes methods for aligning reads (e.g., nucleic acid reads, amino acid reads) to a reference sequence construct, methods for building the reference sequence construct, and systems that use the alignment methods and constructs to produce sequences. The method is scalable, and can be used to align millions of reads to a construct thousands of bases or amino acids long. The invention additionally includes methods for identifying a disease or a genotype based upon alignment of nucleic acid reads to a location in the construct. 02-26-2015
20150056613 METHODS AND SYSTEMS FOR DETECTING SEQUENCE VARIANTS - The invention provides methods for identifying rare variants near a structural variation in a genetic sequence, for example, in a nucleic acid sample taken from a subject. The invention additionally includes methods for aligning reads (e.g., nucleic acid reads) to a reference sequence construct accounting for the structural variation, methods for building a reference sequence construct accounting for the structural variation or the structural variation and the rare variant, and systems that use the alignment methods to identify rare variants. The method is scalable, and can be used to align millions of reads to a construct thousands of bases long, or longer.
We expect those to be just the appetizer on what to come, because the company is still trying to hire more patent writers.
Position
At Seven Bridges our dedicated R&D; and scientific teams are expanding the frontiers of bioinformatics, developing cutting edge tools and methods based on novel insights at the intersection of math, computer science, and biology. Immersed in the latest scientific breakthrough and publications, you would play a crucial role in keeping this innovation engine running by securing legal recognition for our inventions and building a bulwark of intellectual property to match our established competitors.
As a patent technical writer you would be involved in all stages of the patent process at Seven Bridges. You would work with our product and engineering teams to identify patentable inventions and apply your knowledge of the patent system, our product, and our competitive environment to determine which inventions to seek patent protection for. You would then draft disclosures describing inventions with the assistance of their inventors, review patent applications drafted by patent counsel on the basis of these disclosures, and assist patent counsel and internal decision-makers in making strategic judgments throughout the application process.
An ideal candidate would have
bachelor’s degree
experience in math, computer science, or computational biology
ability to produce professional documents
strong organizational skills
And we also think that
2+ years experience working with the patent system, ideally working on drafting and patent prosecution, is a plus
technical writing experience is a plus
familiarity with computer science and software engineering is a plus
We are also looking for a strong cultural fit, as we value our team more than anything else and we like to learn from each other and share the knowledge. In return, we offer a relaxed and open work environment, great work conditions, and first-hand involvement in one of the most exciting fields of science that is laying down the foundations of modern personalized medicine.
If you would like to help us advance genomics at our leading, well-funded, and dynamic startup, please apply here. Thank you for your interest in Seven Bridges Genomics, however, due to the high volume of submissions we will only contact those candidates whose skills and qualifications most closely suit our needs.