Note: These tutorials are incomplete. More complete versions are being made available for our members. Sign up for free.

The World of Mapping Programs
The number of available mapping programs <A href=http://wwwdev.ebi.ac.uk/fg/hts_mappers/>expanded rapidly in recent years</A> due to the advent of next-generation sequencing (NGS). A review paper by <A href=http://www.biostat.jhsph.edu/bstcourse/bio638/readings/li_homer_shortreadaln_review.pdf>Li and Homer</A> provides good introduction to several NGS-related algorithms.
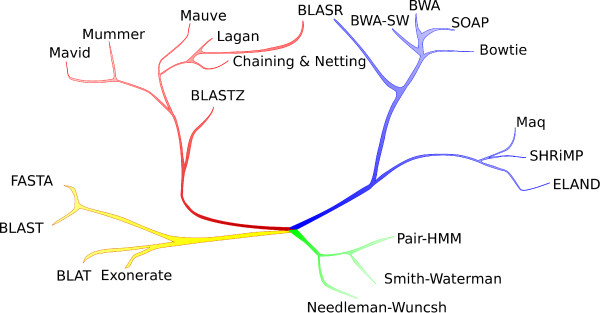
Many of those programs overlap with each other and the set of addressed biological problems and core mathematical concepts did not increase as rapidly as the number of programs. The above image, borrowed from <A href=http://www.biomedcentral.com/content/pdf/1471-2105-13-238.pdf>Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory</A> by Mark J Chaisson and Glenn Tesler, presents the evolutionary relationship between various mapping programs. The programs within the same branch of the tree are conceptually similar. In fact, the core set of mathematical ideas implemented by different branches is even fewer.
