Note: These tutorials are incomplete. More complete versions are being made available for our members. Sign up for free.

The Genome Assembly Problem
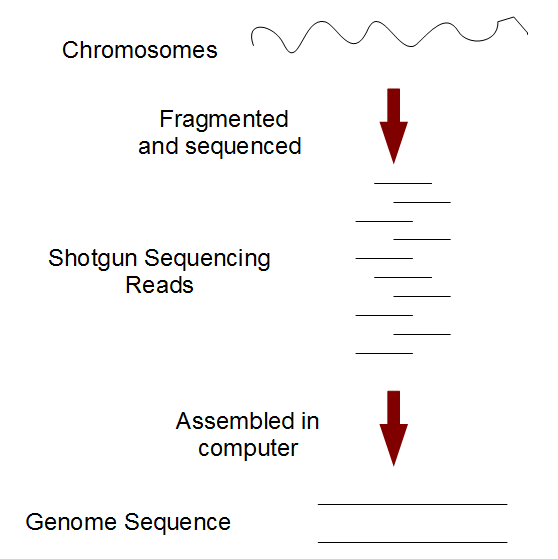
A typical eukaryotic chromosome is millions of nucleotides long. No sequencing technology of present time has the ability to decode the entire sequence in one shot. Therefore, genome sequencing requires additional strategies beyond the use of sequencing instruments. In a shotgun approach, the chromosome is chemically parsed into many small fragments and each fragment is decoded by the sequencing instruments. Subsequently, a specialized computer program (genome assembler) merges all small pieces together to computationally rebuild the genome sequence.
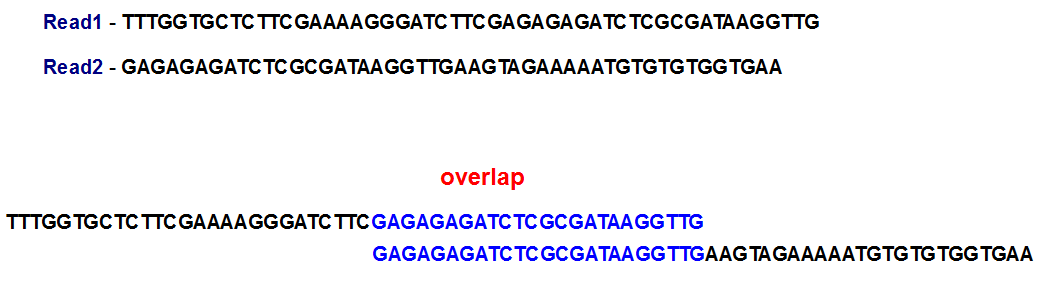
How does a genome assembler work? In traditional overlap-consensus-layout method used for assembling Sanger reads, the assembler identifies overlaps between various long reads. Based on those overlaps, it subsequently merges the read fragments into longer sequences.
